9ES7
 
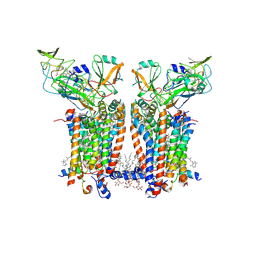 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with water molecules at 1.94 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
4UVS
 
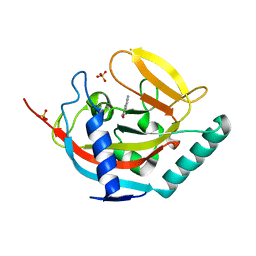 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- pentyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-pentylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4W8D
 
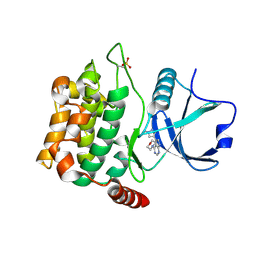 | | Crystal structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06454589). | | Descriptor: | 5-(1-methyl-1H-pyrazol-4-yl)-4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidine, Serine/threonine-protein kinase 24 36 kDa subunit | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
9IHS
 
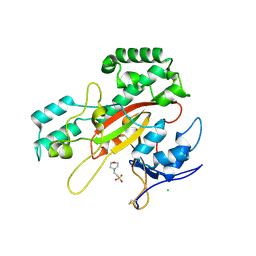 | | Microbial transglutaminase mutant - D3C/G283C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Suzuki, M, Date, M, Kashiwagi, T, Takahashi, K, Nakamura, A, Tanokura, M, Suzuki, E, Yokoyama, K. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Random mutagenesis and disulfide bond formation improved thermostability in microbial transglutaminase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
4V9R
 
 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
3JCM
 
 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
7LJH
 
 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Zn+2 bound | | Descriptor: | Poly(Aspartic acid) hydrolase, ZINC ION | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
7LJI
 
 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Gd+3 bound | | Descriptor: | GADOLINIUM ION, Poly(Aspartic acid) hydrolase | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
3K8V
 
 | |
7LML
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 6-iodanyl-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
3JRR
 
 | | Crystal structure of the ligand binding suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3 | | Authors: | Chan, J, Ishiyama, N, Ikura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 1.9 angstrom crystal structure of the suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor
To be Published
|
|
4WE2
 
 | | Donor strand complemented FaeG of F4ab fimbriae | | Descriptor: | K88 fimbrial protein AB | | Authors: | Moonens, K, Van den Broeck, I, De Kerpel, M, Deboeck, F, Raymaekers, H, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Insight into the Carbohydrate Receptor Binding of F4 Fimbriae-producing Enterotoxigenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
9EY5
 
 | |
4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
3JSV
 
 | | Crystal structure of mouse NEMO CoZi in complex with Lys63-linked di-ubiquitin | | Descriptor: | NF-kappa-B essential modulator, Ubiquitin | | Authors: | Yoshikawa, A, Sato, Y, Mimura, H, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-09-11 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin
Febs Lett., 583, 2009
|
|
3K9R
 
 | | X-ray structure of the Rhodanese-like domain of the Alr3790 protein from Anabaena sp. Northeast Structural Genomics Consortium Target NsR437c. | | Descriptor: | Alr3790 protein | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Maglaqui, M, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of the Rhodanese-like domain of the Alr3790 protein from Anabaena sp.
To be Published
|
|
9FOY
 
 | | Ternary complex of a Mycobacterium tuberculosis DNA gyrase core fusion with DNA and the inhibitor AMK32b | | Descriptor: | (1~{R})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, (1~{S})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), ... | | Authors: | Kokot, M, Hrast, M, Feng, L, Mitchenall, L.A, Lawson, D.M, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2024-06-12 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the Mycobacterium tuberculosis DNA gyrase-DNA-NBTI complex
To be published
|
|
9EWK
 
 | | Solvent organization in ultrahigh-resolution protein crystal structure at room temperature | | Descriptor: | Crambin, ETHANOL | | Authors: | Chen, J.C.-H, Gilski, M, Chang, C, Borek, D, Rosenbaum, G, Lavens, A, Otwinowski, Z, Kubicki, M, Dauter, Z, Jaskolski, M, Joachimiak, A. | | Deposit date: | 2024-04-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Solvent organization in the ultrahigh-resolution crystal structure of crambin at room temperature.
Iucrj, 11, 2024
|
|
4UZJ
 
 | |
9EY6
 
 | | Crystal structure of human tyrosinase-related protein 1 (TYRP1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5,6-dihydroxyindole-2-carboxylic acid oxidase, ... | | Authors: | Ng, Y.M, Soler-Lopez, M. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | Interactions of Phenylalanine Derivatives with Human Tyrosinase: Lessons from Experimental and Theoretical tudies.
Chembiochem, 25, 2024
|
|
4V4L
 
 | | Structure of the Drosophila apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK, MAGNESIUM ION | | Authors: | Yuan, S, Topf, M, Akey, C.W, Ludtke, S.J. | | Deposit date: | 2010-10-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the Drosophila apoptosome at 6.9 angstrom resolution
Structure, 19, 2011
|
|
4V4Y
 
 | | Crystal structure of the 70S Thermus thermophilus ribosome with translocated and rotated Shine-Dalgarno Duplex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
9EY7
 
 | |
7LA5
 
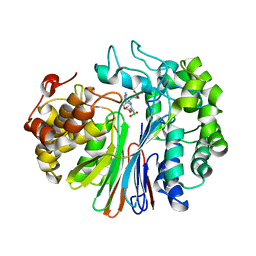 | | Structure of human GGT1 in complex with Lnt1-172 compound. | | Descriptor: | (2R)-4-borono-2-{[(1H-imidazol-4-yl)methyl]amino}butanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Terzyan, S.S, Hanigan, M, Nguen, L. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of human GGT1 in complex with Lnt1-172 compound.
To Be Published
|
|
4V6G
 
 | | Initiation complex of 70S ribosome with two tRNAs and mRNA. | | Descriptor: | 16S RRNA (E.COLI NUMBERING), 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jenner, L.B, Yusupova, G, Yusupov, M. | | Deposit date: | 2009-07-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural aspects of messenger RNA reading frame maintenance by the ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
