5DCK
 
 | |
5CR8
 
 | | Structure of the membrane-binding domain of pneumolysin | | Descriptor: | Pneumolysin | | Authors: | Marshall, J.E, Faraj, B.H.A, Gingras, A.R, Lonnen, R, Sheikh, M.A, El-Mezgueldi, M, Moody, P.C.E, Andrew, P.W, Wallis, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of Pneumolysin at 2.0 angstrom Resolution Reveals the Molecular Packing of the Pre-pore Complex.
Sci Rep, 5, 2015
|
|
5N1R
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4'-Pyrazol-1-ylmethyl-biphenyl-4-sulfonamide | | Descriptor: | 4-[4-(pyrazol-1-ylmethyl)phenyl]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Scozzafava, A, Carta, F. | | Deposit date: | 2017-02-06 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms
Inorg.Chim.Acta., 470, 2018
|
|
5N4H
 
 | | Crystal structure of the D109N mutant of the mouse alpha-Dystroglycan N-terminal region | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dystroglycan | | Authors: | Cassetta, A, Covaceuszach, S, Brancaccio, A, Sciandra, F, Bozzi, M, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The effect of the pathological V72I, D109N and T190M missense mutations on the molecular structure of alpha-dystroglycan.
PLoS ONE, 12, 2017
|
|
5CRZ
 
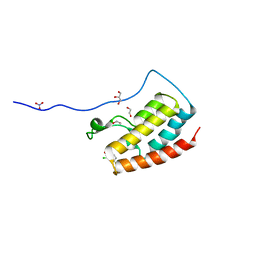 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-4-fluorobenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CS6
 
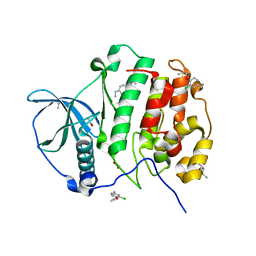 | | Crystal Structure of CK2alpha with Compound 3 bound | | Descriptor: | 1-(3-chloro-4-propoxyphenyl)methanamine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CSY
 
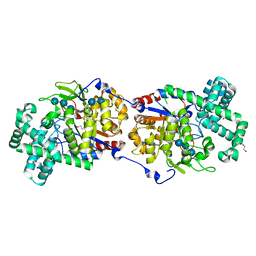 | | Disproportionating enzyme 1 from Arabidopsis - acarbose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5CU6
 
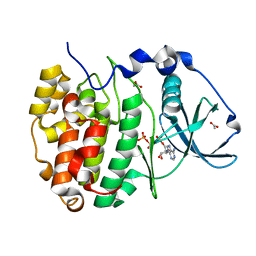 | | Crystal Structure of CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
7CK5
 
 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
5CUM
 
 | |
7CU6
 
 | | lasso peptide C24 mutant - A11V2C | | Descriptor: | lasso peptide C24_A11V2C | | Authors: | Ma, M, Liu, X.H. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7CJW
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a crowded environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
5N6N
 
 | | CRYSTAL STRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX | | Descriptor: | CALCIUM ION, Neutral trehalase, Protein BMH1, ... | | Authors: | Alblova, M, Smidova, A, Obsilova, V, Obsil, T. | | Deposit date: | 2017-02-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5CWW
 
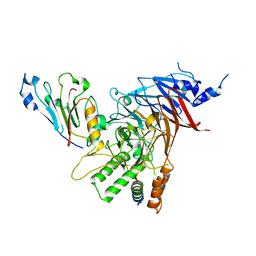 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
7CY6
 
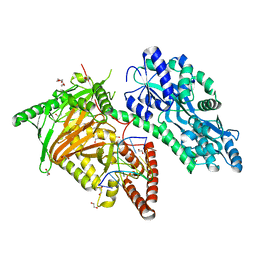 | | Crystal Structure of CMD1 in complex with 5mC-DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
5N7C
 
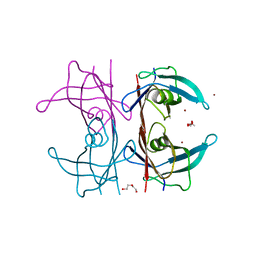 | | Human TTR altered conformation from soaking in CuCl2. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COPPER (II) ION, ... | | Authors: | Ciccone, L, Savko, M, Shepard, W, Stura, E.A. | | Deposit date: | 2017-02-20 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Copper mediated amyloid-beta binding to Transthyretin.
Sci Rep, 8, 2018
|
|
3WJD
 
 | | Crystal structure of mutant nitrobindin F44W/M75L/H76L/Q96C/M148L/H158L (NB5) from Arabidopsis thaliana | | Descriptor: | GLYCEROL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
7CY7
 
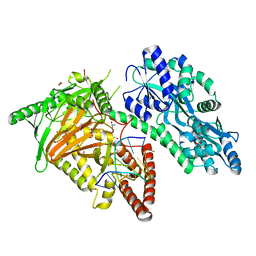 | | Crystal Structure of CMD1 in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7D0F
 
 | | cryo-EM structure of a pre-catalytic group II intron RNP | | Descriptor: | Group II intron-encoded protein LtrA, RNA (738-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
5DG9
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA INSERTING dGMPNPP ACROSS A DNA TEMPLATE CONTAINING 1,N6-ETHENODEOXYADENOSINE LESION | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(EDA)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5CYI
 
 | | CDK2/Cyclin A covalent complex with 6-(cyclohexylmethoxy)-N-(4-(vinylsulfonyl)phenyl)-9H-purin-2-amine (NU6300) | | Descriptor: | 6-(cyclohexylmethoxy)-N-[4-(ethylsulfonyl)phenyl]-9H-purin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Anscombe, E, Meschini, E, Vidal, R.M, Martin, M.P, Staunton, D, Geitmann, M, Danielson, U.H, Stanley, W.A, Wang, L.Z, Reuillon, T, Golding, B.T, Cano, C, Newell, D.R, Noble, M.E.M, Wedge, S.R, Endicott, J.A, Griffin, R.J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Characterization of an Irreversible Inhibitor of CDK2.
Chem.Biol., 22, 2015
|
|
5N88
 
 | | Crystal structure of antibody bound to viral protein | | Descriptor: | PC4 and SFRS1-interacting protein, VH59 antibody | | Authors: | Bao, L, Hannon, C, Cruz-Migoni, A, Ptchelkine, D, Sun, M.-y, Derveni, M, Bunjobpol, W, Chambers, J.S, Simmons, A, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intracellular immunization against HIV infection with an intracellular antibody that mimics HIV integrase binding to the cellular LEDGF protein.
Sci Rep, 7, 2017
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | Descriptor: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
2NV4
 
 | | Crystal structure of UPF0066 protein AF0241 in complex with S-adenosylmethionine. Northeast Structural Genomics Consortium target GR27 | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, UPF0066 protein AF_0241 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
