6XKW
 
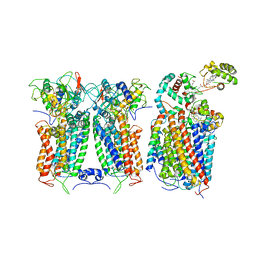 | | R. capsulatus CIII2CIV bipartite super-complex (SC-2A) with CcoH/cy | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
3BIU
 
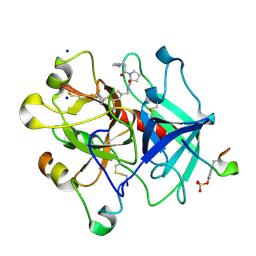 | | Human thrombin-in complex with UB-THR10 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentylamino)ethanoyl)pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin, ... | | Authors: | Gerlach, C, Smolinski, M, Steuber, H, Sotriffer, C.A, Heine, A, Hangauer, D.G, Klebe, G. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Inhibition Profile of a Cyclopentyl and a Cyclohexyl Derivative towards Thrombin: The Same but for Different Reasons
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
3BJL
 
 | |
4Q0S
 
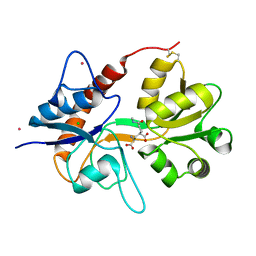
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with ribitol | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), D-ribitol, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
6XX9
 
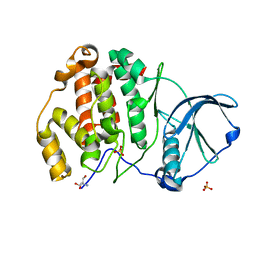 | | Arabidopsis thaliana Casein Kinase 2 (CK2) alpha-1 crystal form III | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Casein kinase II subunit alpha-1, SULFATE ION | | Authors: | Demulder, M, De Veylder, L, Loris, R. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of Arabidopsis thaliana casein kinase 2 alpha 1.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6XY4
 
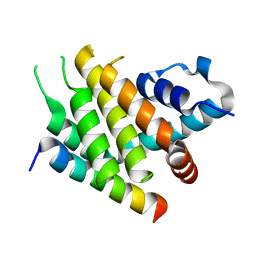 | |
6XJ3
 
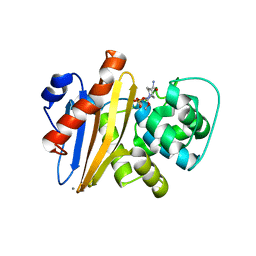 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
6XKX
 
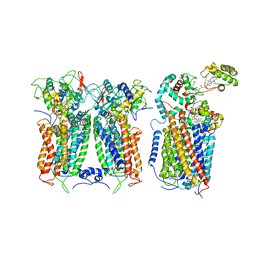 | | R. capsulatus CIII2CIV tripartite super-complex, conformation A (SC-1A) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6YWT
 
 | |
6YVO
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 3.55 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YWU
 
 | | Human OMPD-domain of UMPS (K314AcK) in complex with UMP at 1.1 Angstroms resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
3B43
 
 | | I-band fragment I65-I70 from titin | | Descriptor: | Titin | | Authors: | von Castelmur, E, Marino, M, Labeit, D, Labeit, S, Mayans, O. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4NWC
 
 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | Descriptor: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
6ZQW
 
 | | Cryo-EM structure of immature Spondweni virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, prM | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
6ZRV
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) in Complex with an Inhibitory Nanobody (VHH-s-a93, Nb93) | | Descriptor: | Plasminogen activator inhibitor 1, VHH-s-a93 (Nb93) | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity.
Int J Mol Sci, 21, 2020
|
|
6ZS3
 
 | | Crystal structure of the fifth bromodomain of human protein polybromo-1 in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Protein polybromo-1 | | Authors: | Preuss, F, Joerger, A.C, Wanior, M, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6ZS5
 
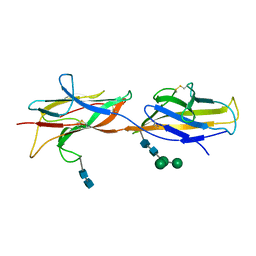 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6ZVI
 
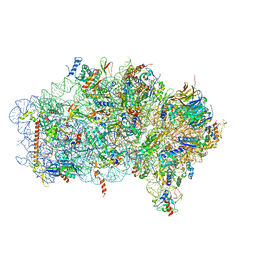 | | Mbf1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
6Z1Z
 
 | | Structure of the anti-CD9 nanobody 4C8 | | Descriptor: | Nanobody 4C8 | | Authors: | Neviani, N, Oosterheert, W, Pearce, N.M, Lutz, M, Kroon-Batenburg, L.M.J, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
6YWN
 
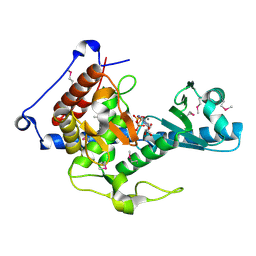 | | CutA in complex with CMPCPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, CALCIUM ION, CutA | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
6YU4
 
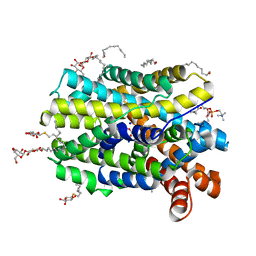 | | Crystal structure of MhsT in complex with L-4F-phenylalanine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4-FLUORO-L-PHENYLALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
3B6C
 
 | |
6ZAV
 
 | | NO-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.19 A resolution (unrestrained, full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
4NGJ
 
 | | Dialyzed HEW lysozyme batch crystallized in 1.0 M RbCl and collected at 100 K | | Descriptor: | CHLORIDE ION, Lysozyme C, RUBIDIUM ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
