2NU3
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2NU2
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
6K05
 
 | | Crystal structure of BRD2(BD1)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
6K04
 
 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
3SGQ
 
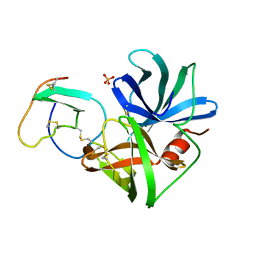 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18,
Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
1CT4
 
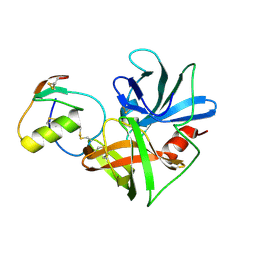 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-VAL18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CSO
 
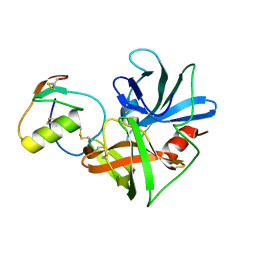 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-ILE18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CT0
 
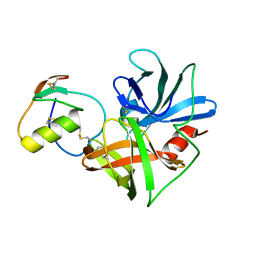 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-SER18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CT2
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-THR18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
2C9A
 
 | | Crystal structure of the MAM-Ig module of receptor protein tyrosine phosphatase mu | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE MU, ... | | Authors: | Aricescu, A.R, Hon, W.C, Siebold, C, Lu, W, Van Der Merwe, P.A, Jones, E.Y. | | Deposit date: | 2005-12-09 | | Release date: | 2006-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Analysis of Receptor Protein Tyrosine Phosphatase Mu-Mediated Cell Adhesion.
Embo J., 25, 2006
|
|
5SVR
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel bound to competitive antagonist A-317491 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-{[(3-phenoxyphenyl)methyl][(1S)-1,2,3,4-tetrahydronaphthalen-1-yl]carbamoyl}benzene-1,2,4-tricarboxylic acid, MAGNESIUM ION, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVJ
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel in the closed, apo state | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVQ
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel bound to competitive antagonist TNP-ATP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, P2X purinoceptor 3, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVM
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel bound to agonist 2-methylthio-ATP in the desensitized state | | Descriptor: | 1,2-ETHANEDIOL, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.093 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVP
 
 | | Anomalous sulfur signal reveals the position of agonist 2-methylthio-ATP bound to the ATP-gated human P2X3 ion channel in the desensitized state | | Descriptor: | 1,2-ETHANEDIOL, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVK
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel in the ATP-bound, open state | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVT
 
 | | Anomalous Cs+ signal reveals the site of Na+ ion entry to the channel pore of the human P2X3 ion channel through the extracellular fenestrations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, P2X purinoceptor 3, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.794 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVL
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel in the ATP-bound, closed (desensitized) state | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-06 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVS
 
 | | Anomalous Mn2+ signal reveals a divalent cation-binding site in the head domain of the ATP-gated human P2X3 ion channel | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.025 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5WP6
 
 | | Cryo-EM structure of a human TRPM4 channel in complex with calcium and decavanadate | | Descriptor: | DECAVANADATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Winkler, P.A, Huang, Y, Sun, W, Du, J, Lu, W. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Electron cryo-microscopy structure of a human TRPM4 channel.
Nature, 552, 2017
|
|
8EQ4
 
 | | Human PAC in nanodisc at pH 4.0 with PI(4,5)P2 diC8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proton-activated chloride channel, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Ruan, Z, Lu, W. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Inhibition of the proton-activated chloride channel PAC by PIP 2.
Elife, 12, 2023
|
|
8FBL
 
 | | Human PAC in nanodisc at pH 4.0 with PI(4,5)P2 diC8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proton-activated chloride channel, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Ruan, Z, Lu, W. | | Deposit date: | 2022-11-29 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Inhibition of the proton-activated chloride channel PAC by PIP 2.
Elife, 12, 2023
|
|
1ZMM
 
 | |
1ZMP
 
 | | Crystal structure of human defensin-5 | | Descriptor: | CHLORIDE ION, Defensin 5, GLYCEROL, ... | | Authors: | Lubkowski, J, Szyk, A, Lu, W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of human {alpha}-defensins HNP4, HD5, and HD6.
Protein Sci., 15, 2006
|
|
