5JJU
 
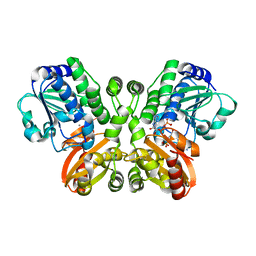 | | Crystal structure of Rv2837c complexed with 5'-pApA and 5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA (5'-R(P*AP*A)-3'), ... | | Authors: | Wang, F, He, Q, Liu, S, Gu, L. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
3L6Y
 
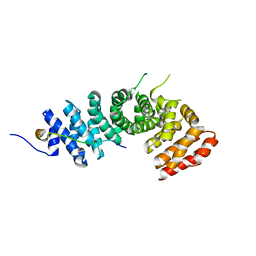 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3UHM
 
 | |
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
8HPF
 
 | |
8HQ7
 
 | |
8HPU
 
 | |
8HP9
 
 | |
8HPV
 
 | |
8HPQ
 
 | |
4WXO
 
 | |
4WXW
 
 | |
7SLP
 
 | | Cryo-EM structure of 7SK core RNP with linear RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Linear 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
7SLQ
 
 | | Cryo-EM structure of 7SK core RNP with circular RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Minimal circular 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
4WHW
 
 | | Direct photocapture of bromodomains using tropolone chemical probes | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-{1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazol-5-yl}cyclohepta-2,4,6-trien-1-one, Bromodomain-containing protein 4 | | Authors: | Hett, E.C, Piatnitski Chekler, E.L, Basak, A, Bonin, P.D, Denny, R.A, Flick, A.C, Geoghegan, K.F, Liu, S, Pletcher, M.T, Robinson, R.P, Sahasrabudhe, P, Salter, S, Stock, I.A, Jones, L.H. | | Deposit date: | 2014-09-23 | | Release date: | 2015-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Direct photocapture of bromodomains using tropolone chemical probes
To Be Published
|
|
8HFC
 
 | | Cryo-EM structure of yeast Erf2/Erf4 complex | | Descriptor: | PALMITIC ACID, Palmitoyltransferase ERF2, Ras modification protein ERF4, ... | | Authors: | Wu, J, Hu, Q, Zhang, Y, Yang, A, Liu, S. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HF3
 
 | | Cryo-EM structure of human ZDHHC9/GCP16 complex | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Golgin subfamily A member 7, PALMITIC ACID, ... | | Authors: | Wu, J, Hu, Q, Zhang, Y, Liu, S, Yang, A. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4WXM
 
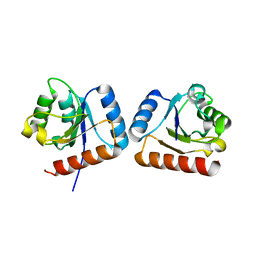 | | FleQ REC domain from Pseudomonas aeruginosa PAO1 | | Descriptor: | Transcriptional regulator FleQ | | Authors: | Su, T, Liu, S, Gu, L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The REC domain mediated dimerization is critical for FleQ from Pseudomonas aeruginosa to function as a c-di-GMP receptor and flagella gene regulator
J.Struct.Biol., 192, 2015
|
|
6OIS
 
 | | CryoEM structure of Arabidopsis DR complex (DMS3-RDM1) | | Descriptor: | Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | Authors: | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
6WVG
 
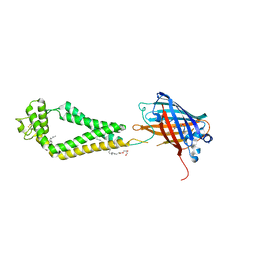 | | human CD53 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein, ... | | Authors: | Yang, Y, Liu, S, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Open conformation of tetraspanins shapes interaction partner networks on cell membranes.
Embo J., 39, 2020
|
|
4ROW
 
 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
4ROV
 
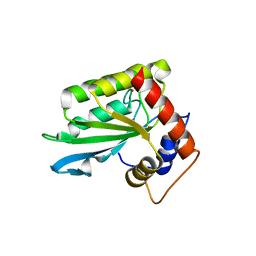 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
