7TIU
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB46 | | Descriptor: | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIZ
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-63 | | Descriptor: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[3-(trifluoromethyl)phenyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIA
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-14 | | Descriptor: | 3C-like proteinase nsp5, THIOCYANATE ION, benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIX
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB56 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TJ0
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
6AGK
 
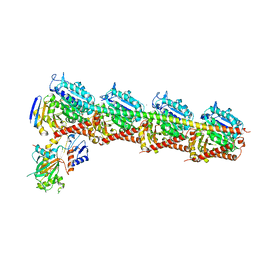 | | The structure of CH-II-77-tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, H, Arnst, K, Wang, Y, Miller, D, Li, W. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
6KTR
 
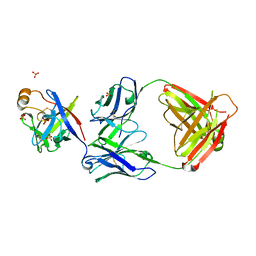 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
3SWH
 
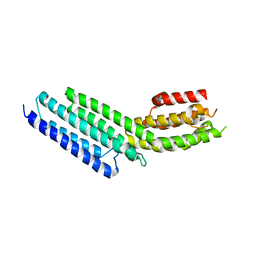 | | Munc13-1, MUN domain, C-terminal module | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Li, W. | | Deposit date: | 2011-07-13 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of a Munc13 C-terminal Module Exhibits a Remarkable Similarity to Vesicle Tethering Factors.
Structure, 19, 2011
|
|
4EL1
 
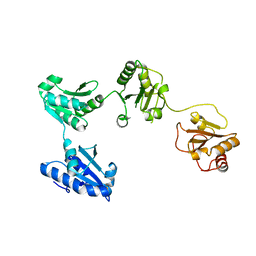 | | Crystal structure of oxidized hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4EKZ
 
 | | Crystal structure of reduced hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4LAM
 
 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in complex with 5-carboxyl-uracil | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
3UA0
 
 | | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease | | Descriptor: | Fibroin heavy chain | | Authors: | He, Y.-X, Zhang, N.-N, Chen, B.-Y, Li, W.-F, Chen, Y.-X, Zhou, C.-Z. | | Deposit date: | 2011-10-20 | | Release date: | 2012-03-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N-Terminal Domain of Bombyx mori Fibroin Mediates the Assembly of Silk in Response to pH Decrease.
J.Mol.Biol., 418, 2012
|
|
4LAN
 
 | | Crystal structure of Cordyceps militaris IDCase H195A mutant | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAO
 
 | | Crystal structure of Cordyceps militaris IDCase H195A mutant (Zn) | | Descriptor: | Cordyceps militaris IDCase, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
1G7P
 
 | | CRYSTAL STRUCTURE OF MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND YEAST ALPHA-GLUCOSIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-GLUCOSIDASE P1, ... | | Authors: | Apostolopoulos, V, Yu, M, Corper, A.L, Li, W, McKenzie, I.F, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-11-13 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a non-canonical high affinity peptide complexed with MHC class I: a novel use of alternative anchors.
J.Mol.Biol., 318, 2002
|
|
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
4DY0
 
 | | Crystal structure of native protease nexin-1 with heparin | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, GLYCEROL, Glia-derived nexin, ... | | Authors: | Huntington, J.A, Li, W. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of protease nexin-1 in complex with heparin and thrombin suggest a 2-step recognition mechanism.
Blood, 120, 2012
|
|
1Z6T
 
 | | Structure of the apoptotic protease-activating factor 1 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apoptotic protease activating factor 1 | | Authors: | Riedl, S.J, Li, W, Chao, Y, Schwarzenbacher, R, Shi, Y. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the apoptotic protease-activating factor 1 bound to ADP
Nature, 434, 2005
|
|
2P3N
 
 | | Thermotoga maritima IMPase TM1415 | | Descriptor: | Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Stieglitz, K.A, Roberts, M.F, Li, W, Stec, B. | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
Febs J., 274, 2007
|
|
2P3V
 
 | | Thermotoga maritima IMPase TM1415 | | Descriptor: | Inositol-1-monophosphatase, S,R MESO-TARTARIC ACID | | Authors: | Stieglitz, K.A, Roberts, M.F, Li, W, Stec, B. | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
Febs J., 274, 2007
|
|
6BFU
 
 | | Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein, ... | | Authors: | Xiong, X, Tortorici, M.A, Snijder, S, Yoshioka, C, Walls, A.C, Li, W, Seattle Structural Genomics Center for Infectious Disease (SSGCID), McGuire, A.T, Rey, F.A, Bosch, B.J, Veesler, D. | | Deposit date: | 2017-10-27 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Glycan Shield and Fusion Activation of a Deltacoronavirus Spike Glycoprotein Fine-Tuned for Enteric Infections.
J. Virol., 92, 2018
|
|
5H7O
 
 | | Crystal structure of DJ-101 in complex with tubulin protein | | Descriptor: | 2-(1H-indol-4-yl)-4-(3,4,5-trimethoxyphenyl)-1H-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Arnst, K, Wang, Y, Hwang, D.-J, Xue, Y, Costello, T, Hamilton, D, Chen, Q, Yang, J, Park, F, Dalton, J.T, Miller, D.D, Li, W. | | Deposit date: | 2016-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Potent, Metabolically Stable Tubulin Inhibitor Targets the Colchicine Binding Site and Overcomes Taxane Resistance.
Cancer Res., 78, 2018
|
|
3RN4
 
 | | Crystal structure of iron-substituted Sod2 from Saccharomyces cerevisiae | | Descriptor: | FE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Kang, Y, He, Y.-X, Cheng, W, Zhou, C.-Z, Li, W.-F. | | Deposit date: | 2011-04-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structures of native and Fe-substituted SOD2 from Saccharomyces cerevisiae
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4QAU
 
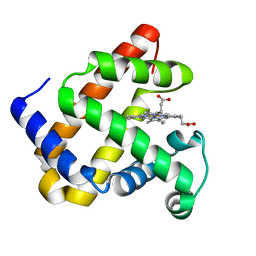 | | Crystal structure of F43Y mutant of sperm whale myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y, Tan, X, Li, W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | A Novel Tyrosine-Heme C-O Covalent Linkage in F43Y Myoglobin: A New Post-translational Modification of Heme Proteins
Chembiochem, 16, 2015
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
