5A9K
 
 | | Structural basis for DNA strand separation by a hexameric replicative helicase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, REPLICATION PROTEIN E1 | | Authors: | Chaban, Y, Stead, J.A, Ryzhenkova, K, Whelan, F, Lamber, K, Antson, F, Sanders, C.M, Orlova, E.V. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Structural Basis for DNA Strand Separation by a Hexameric Replicative Helicase.
Nucleic Acids Res., 43, 2015
|
|
7A08
 
 | | CryoEM Structure of cGAS Nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Michalski, S, de Oliveira Mann, C.C, Witte, G, Bartho, J, Lammens, K, Hopfner, K.P. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-23 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for sequestration and autoinhibition of cGAS by chromatin.
Nature, 587, 2020
|
|
2QFD
 
 | | Crystal structure of the regulatory domain of human RIG-I with bound Hg | | Descriptor: | MERCURY (II) ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Cui, S, Lammens, A, Lammens, K, Hopfner, K.P. | | Deposit date: | 2007-06-27 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I.
Mol.Cell, 29, 2008
|
|
4FCX
 
 | | S.pombe Mre11 apoenzym | | Descriptor: | DNA repair protein rad32, MANGANESE (II) ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FBW
 
 | |
5DAC
 
 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with DNA | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
6RR9
 
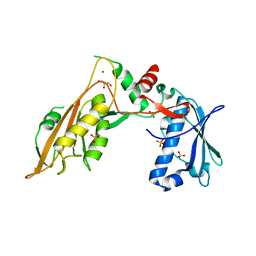 | | DNA/RNA binding protein | | Descriptor: | GLYCEROL, SULFATE ION, Schlafen family member 5, ... | | Authors: | Huber, E, Lammens, K. | | Deposit date: | 2019-05-17 | | Release date: | 2020-07-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.432 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 2022
|
|
6S6V
 
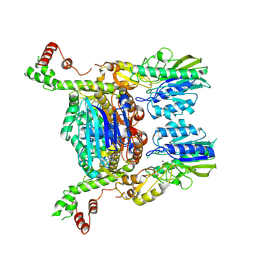 | | Resting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ATPgS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Nuclease SbcCD subunit C, ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-03 | | Release date: | 2019-09-04 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
4I1R
 
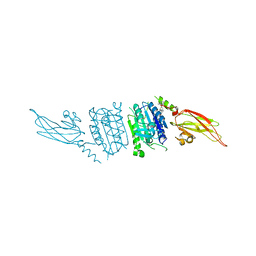 | | Human MALT1 (caspase-IG3) in complex with thioridazine | | Descriptor: | 10-{2-[(2S)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Schlauderer, F, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analysis of Phenothiazine Derivatives as Allosteric Inhibitors of the MALT1 Paracaspase.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4I51
 
 | | Methyltransferase domain of HUMAN EUCHROMATIC HISTONE METHYLTRANSFERASE 1, mutant Y1211A | | Descriptor: | GLYCEROL, H3K9 NE-ALLYL PEPTIDE, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Islam, K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Lou, M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-11-28 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Defining efficient enzyme-cofactor pairs for bioorthogonal profiling of protein methylation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4I1P
 
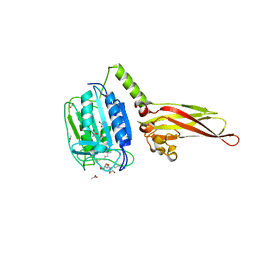 | |
6S85
 
 | | Cutting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), DNA (32-MER), ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
6GPG
 
 | | Structure of the RIG-I Singleton-Merten syndrome variant C268F | | Descriptor: | MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*CP*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*CP*G)-3'), ... | | Authors: | Laessig, C, Lammens, K, Hopfner, K.-P. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Unified mechanisms for self-RNA recognition by RIG-I Singleton-Merten syndrome variants.
Elife, 7, 2018
|
|
6GK2
 
 | | Helical reconstruction of BCL10 CARD and MALT1 DEATH DOMAIN complex | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Schlauderer, F, Desfosses, A, Gutsche, I, Hopfner, K.P, Lammens, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-31 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture and regulation of BCL10-MALT1 filaments.
Nat Commun, 9, 2018
|
|
5DA9
 
 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with the Rad50-binding domain of Mre11 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Putative double-strand break protein, ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
7ZR1
 
 | | Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS (composite structure) | | Descriptor: | DH domain-containing protein, Double-strand break repair protein, FHA domain-containing protein, ... | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-05-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
8BAH
 
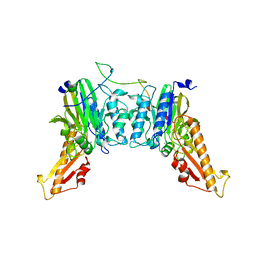 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
2QFB
 
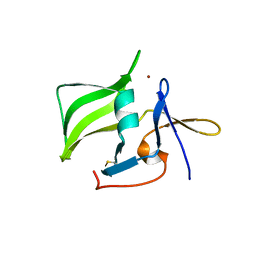 | | Crystal structure of the regulatory domain of human RIG-I with bound Zn | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, ZINC ION | | Authors: | Cui, S, Lammens, A, Lammens, K, Hopfner, K.P. | | Deposit date: | 2007-06-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I.
Mol.Cell, 29, 2008
|
|
6D08
 
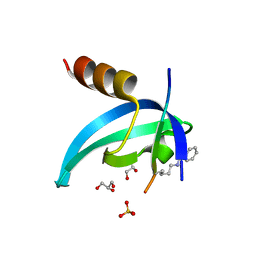 | | Crystal structure of an engineered bump-hole complex of mutant human chromobox homolog 1 (CBX1) with H3K9bn peptide | | Descriptor: | Chromobox protein homolog 1, GLYCEROL, Histone H3.1, ... | | Authors: | Arora, S, Horne, W.S, Islam, K. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Methyllysine Writers and Readers for Allele-Specific Regulation of Protein-Protein Interactions.
J.Am.Chem.Soc., 141, 2019
|
|
3QF7
 
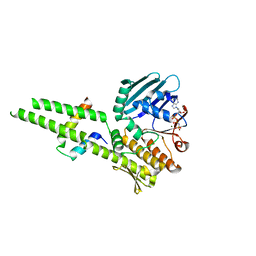 | |
6D07
 
 | |
7YZP
 
 | | Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and a DNA hairpin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA hairpin (59-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7YZO
 
 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7Z03
 
 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (39-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7Z8S
 
 | | Mot1:TBP:DNA - post hydrolysis state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
