2WXO
 
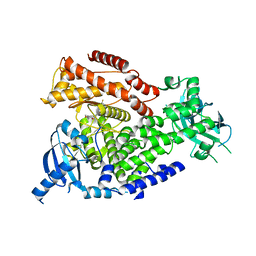 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with AS5. | | Descriptor: | N-(3-{[(1Z)-3,5-DIMETHOXYCYCLOHEXA-2,4-DIEN-1-YLIDENE]AMINO}QUINOXALIN-2-YL)-4-FLUOROBENZENESULFONAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXJ
 
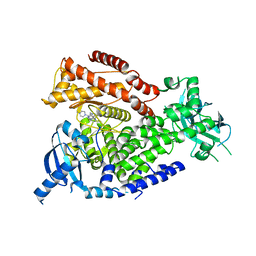 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with INK654. | | Descriptor: | N-[6-(4-amino-1-{[2-(4-methylpiperazin-1-yl)quinolin-3-yl]methyl}-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzothiazol-2-yl]acetamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
8TA5
 
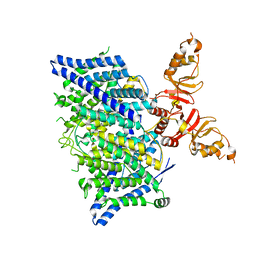 | | Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA4
 
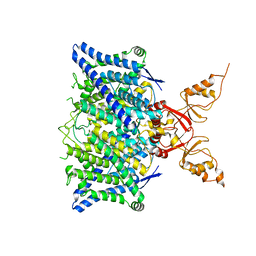 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA3
 
 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA2
 
 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42 | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA6
 
 | | Cryo-EM structure of the human CLC-2 chloride channel C-terminal domain | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
2OUS
 
 | | crystal structure of PDE10A2 mutant D674A | | Descriptor: | MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUQ
 
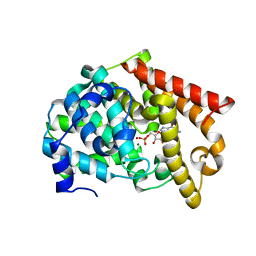 | | crystal structure of PDE10A2 in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4RXE
 
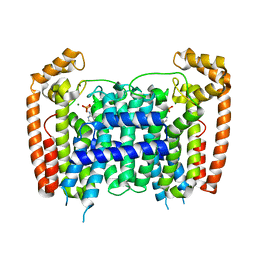 | |
4AOT
 
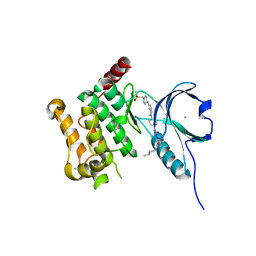 | | Crystal Structure of Human Serine Threonine Kinase-10 (LOK) Bound to GW830263A | | Descriptor: | 1-(4-{methyl[2-({4-[(methylsulfonyl)methyl]phenyl}amino)pyrimidin-4-yl]amino}phenyl)-3-{3-[(4-methylpiperazin-1-yl)carbonyl]phenyl}urea, CHLORIDE ION, Serine/threonine-protein kinase 10 | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, Canning, P, von Delft, F, Yue, W, Liu, Y, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of Human Serine Threonine Kinase-10 (Lok) Bound to Gw830263A
To be Published
|
|
8K7O
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design protein -T03
To Be Published
|
|
4RXC
 
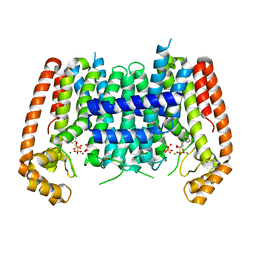 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Homorisedronate BPH-6 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [1-hydroxy-3-(pyridin-3-yl)propane-1,1-diyl]bis(phosphonic acid) | | Authors: | Cao, R, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Farnesyl diphosphate synthase inhibitors with unique ligand-binding geometries.
ACS Med Chem Lett, 6, 2015
|
|
8S9P
 
 | | 1:1:1 agrin/LRP4/MuSK complex | | Descriptor: | Agrin, Low-density lipoprotein receptor-related protein 4, Muscle, ... | | Authors: | Xie, T, Xu, G.J, Liu, Y, Quade, B, Lin, W.C, Bai, X.C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the assembly of the agrin/LRP4/MuSK signaling complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4ZRS
 
 | | Crystal structure of a cloned feruloyl esterase from a soil metagenomic library | | Descriptor: | Esterase, GLYCEROL | | Authors: | Xie, W, Chen, R, Cao, L, Liu, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing the Thermostability of Feruloyl Esterase EstF27 by Directed Evolution and the Underlying Structural Basis
J.Agric.Food Chem., 63, 2015
|
|
8TGV
 
 | | CryoEM structure of Fab HC84.26-HCV E2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | Authors: | Shahid, S, Liqun, J, LIu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | CryoEM structure of Fab HC84.26-HCV E2 complex
To Be Published
|
|
8TGZ
 
 | | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8THZ
 
 | | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CBH-7 Heavy chain, ... | | Authors: | Shahid, S, Jiang, L, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8TXQ
 
 | | CryoEM structure of non-neutralizing antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | CBH-4B Heavy chain, CBH-4B Light chain, envelope glycoprotein E2 | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of non-neutralizing antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8TZY
 
 | | CryoEM structure of non-neutralizing bivalent antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | CBH4B Heavy chain, CBH4B Light chain, envelope glycoprotein E2 | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-08-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM structure of non-neutralizing bivalent antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
4A5V
 
 | | Solution structure ensemble of the two N-terminal apple domains (residues 58-231) of Toxoplasma gondii microneme protein 4 | | Descriptor: | MICRONEMAL PROTEIN 4 | | Authors: | Marchant, J, Cowper, B, Liu, Y, Lai, L, Pinzan, C, Marq, J.B, Friedrich, N, Sawmynaden, K, Chai, W, Childs, R.A, Saouros, S, Simpson, P, Barreira, M.C.R, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Galactose Recognition by the Apicomplexan Parasite Toxoplasma Gondii.
J.Biol.Chem., 287, 2012
|
|
1GIS
 
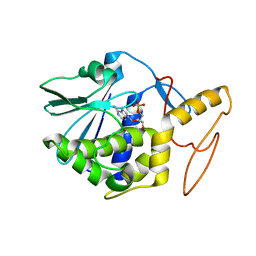 | | A TRICHOSANTHIN(TCS) MUTANT(E85Q) COMPLEX STRUCTURE WITH 2'-DEOXY-ADENOSIN-5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Guo, Q, Liu, Y, Dong, Y, Rao, Z. | | Deposit date: | 2001-03-15 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate binding and catalysis in trichosanthin occur in different sites as revealed by the complex structures of several E85 mutants.
Protein Eng., 16, 2003
|
|
3QWY
 
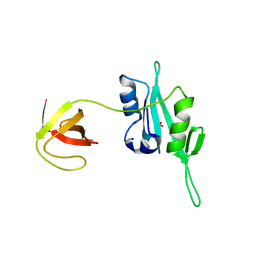 | | CED-2 | | Descriptor: | Cell death abnormality protein 2, GLYCEROL, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y, Sun, D, Hu, Y, Liu, Y.F. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
1NXK
 
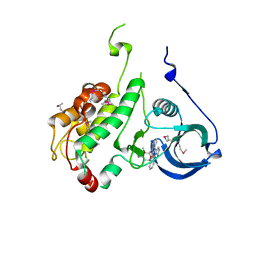 | | Crystal structure of staurosporine bound to MAP KAP kinase 2 | | Descriptor: | MAP kinase-activated protein kinase 2, STAUROSPORINE, SULFATE ION | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Czerwinski, R.M, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-10 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
3TI6
 
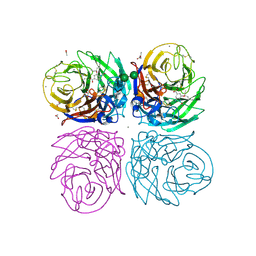 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
