1UNG
 
 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | 6-PHENYL[5H]PYRROLO[2,3-B]PYRAZINE, CELL DIVISION PROTEIN KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNH
 
 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
2B4L
 
 | | Crystal structure of the binding protein OpuAC in complex with glycine betaine | | Descriptor: | 1,2-ETHANEDIOL, Glycine betaine-binding protein, TRIMETHYL GLYCINE | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
1UYM
 
 | | Human Hsp90-beta with PU3 (9-Butyl-8(3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine) | | Descriptor: | 9-BUTYL-8-(3,4,5-TRIMETHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-BETA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UYF
 
 | | Human Hsp90-alpha with 8-(2-chloro-3,4,5-trimethoxy-benzyl)-2-fluoro-9-pent-4-ylnyl-9H-purin-6-ylamine | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
4X5T
 
 | | alpha 1 glycine receptor transmembrane structure fused to the extracellular domain of GLIC | | Descriptor: | ACETATE ION, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Sauguet, L, Corringer, P.J, Huon, C, Delarue, M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Allosteric and hyperekplexic mutant phenotypes investigated on an alpha 1 glycine receptor transmembrane structure.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X6A
 
 | | Crystal structure of yeast RNA polymerase II encountering oxidative Cyclopurine DNA lesions | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8DYP
 
 | |
1ID4
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157Q) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-03 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
2QGS
 
 | | Crystal structure of SE1688 protein from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR89 | | Descriptor: | Protein SE1688, ZINC ION | | Authors: | Forouhar, F, Su, M, Benach, J, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Owen, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of SE1688 protein from Staphylococcus epidermidis.
To be Published
|
|
1YBX
 
 | | Conserved hypothetical protein Cth-383 from Clostridium thermocellum | | Descriptor: | Conserved hypothetical protein, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Chang, J, Zhao, M, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Ljundahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved hypothetical protein Cth-383 from Clostridium thermocellum
To be published
|
|
1ZK5
 
 | | Escherichia coli F17fG lectin domain complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17G adhesin subunit | | Authors: | Buts, L, Wellens, A, Van Molle, I, De Genst, E, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-02 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of natural variation in bacterial F17G adhesins on crystallization behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y80
 
 | | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica | | Descriptor: | CO-5-METHOXYBENZIMIDAZOLYLCOBAMIDE, Predicted cobalamin binding protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Z.-J, Fu, Z.-Q, Tempel, W, Das, A, Habel, J, Zhou, W, Chang, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Ljungdahl, L, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica
To be published
|
|
1ZKD
 
 | | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58 | | Descriptor: | DUF185 | | Authors: | Kuzin, A.P, Yong, W, Vorobiev, S.M, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58
To be Published
|
|
1YDP
 
 | | 1.9A crystal structure of HLA-G | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Clements, C.S, Kjer-nielsen, L, Kostenko, L, Hoare, H.L, Dunstone, M.A, Moses, E, Freed, K, Brooks, A.G, Rossjohn, J, Mccluskey, J. | | Deposit date: | 2004-12-25 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of HLA-G: A nonclassical MHC class I molecule expressed at the fetal-maternal interface
PROC.NATL.ACAD.SCI.USA, 102, 2005
|
|
8OSL
 
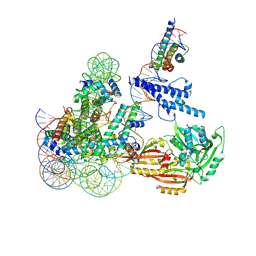 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTS
 
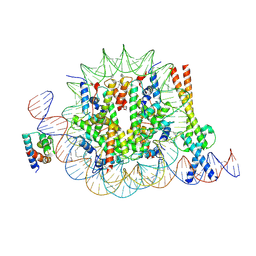 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OTT
 
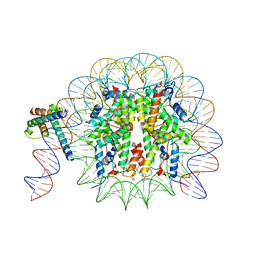 | | MYC-MAX bound to a nucleosome at SHL+5.8 | | Descriptor: | DNA (144-MER), Histone H2A type 1-B/E, Histone H2A type 1-K, ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Kater, L, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
1Y6J
 
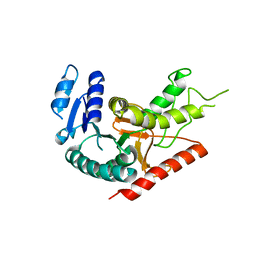 | | L-Lactate Dehydrogenase from Clostridium Thermocellum Cth-1135 | | Descriptor: | L-lactate dehydrogenase | | Authors: | Chen, L, Yang, H, Kataeva, I, Chen, L.R, Tempel, W, Lee, D, Habel, J, Zhou, W, Lin, D, Ljungdahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-06 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | L-Lactate Dehydrogenase from Clostridium Thermocellum Cth-1135
To be Published
|
|
1Z8U
 
 | | Crystal structure of oxidized alpha hemoglobin bound to AHSP | | Descriptor: | Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Feng, L, Zhou, S, Gu, L, Gell, D.A, Mackay, J.P, Weiss, M.J, Gow, A.J, Shi, Y. | | Deposit date: | 2005-03-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of oxidized alpha-haemoglobin bound to AHSP reveals a protective mechanism for haem.
Nature, 435, 2005
|
|
1YCY
 
 | | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein | | Authors: | Huang, L, Liu, Z.-J, Lee, D, Tempel, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus
To be published
|
|
2QZ4
 
 | | Human paraplegin, AAA domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Paraplegin | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, J, Sagemark, C, Sundstrom, M, Thorsell, A.G, Tresauges, L, Van Den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of the ATPase Domain of the Human AAA+ Protein Paraplegin/SPG7.
Plos One, 4, 2009
|
|
2QZG
 
 | | Crystal structure of unknown function protein MMP1188 | | Descriptor: | Conserved uncharacterized archaeal protein | | Authors: | Chang, C, Perez, V, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of MMP1188, unknown function protein.
To be Published
|
|
