5T04
 
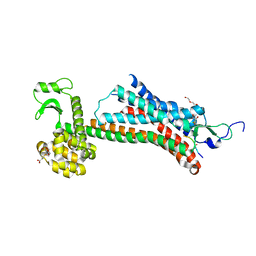 | | STRUCTURE OF CONSTITUTIVELY ACTIVE NEUROTENSIN RECEPTOR | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ARG-ARG-PRO-TYR-ILE-LEU, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B, Botos, I, Grisshammer, R. | | Deposit date: | 2016-08-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and dynamics of a constitutively active neurotensin receptor.
Sci Rep, 6, 2016
|
|
3F62
 
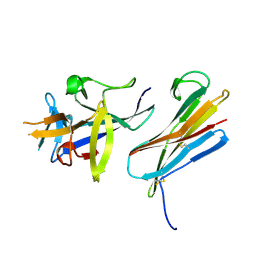 | |
9CIB
 
 | | CryoEM Structure of HCA2 DREADD Gi1 in complex with FCH-2296413 (Local Refinement) | | Descriptor: | (3M,4aR,5aR)-3-(1H-tetrazol-5-yl)-4,4a,5,5a-tetrahydro-1H-cyclopropa[4,5]cyclopenta[1,2-c]pyrazole, Hydroxycarboxylic acid receptor 2 | | Authors: | Krumm, B.E, Kang, H.J, Diberto, J.F, Kapolka, N.J, Gumpper, R.H, Olsen, R.H.J, Huang, X.P, Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-07-03 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided design of a peripherally restricted chemogenetic system.
Cell, 187, 2024
|
|
4XFU
 
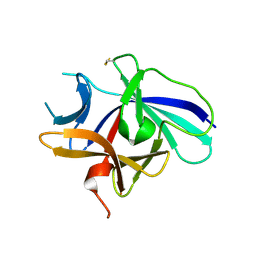 | | Structure of IL-18 SER Mutant V | | Descriptor: | Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5CZ3
 
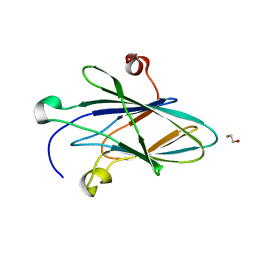 | | Crystal Structure of Myxoma Virus M64 | | Descriptor: | BETA-MERCAPTOETHANOL, M64R | | Authors: | Krumm, B.E, Meng, X, Li, Y, Xiang, Y, Deng, J. | | Deposit date: | 2015-07-31 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CYW
 
 | | Crystal Structure of Vaccinia Virus C7 | | Descriptor: | GLYCEROL, Interferon antagonist C7 | | Authors: | Krumm, B.E, Meng, X, Li, Y, Xiang, Y, Deng, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4EKX
 
 | |
4EEE
 
 | |
4XFT
 
 | | Structure of IL-18 SER Mutant III | | Descriptor: | DIMETHYL SULFOXIDE, Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XFS
 
 | | Structure of IL-18 SER Mutant I | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Interleukin-18 | | Authors: | Krumm, B.E, Meng, X, Xiang, Y, Deng, J. | | Deposit date: | 2014-12-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallization of interleukin-18 for structure-based inhibitor design.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
4XEE
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-23 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
8UTD
 
 | | CryoEM Structure of HCA2-Gi1 in complex with MK-1903 | | Descriptor: | (4aR,5aR)-4,4a,5,5a-tetrahydro-1H-cyclopropa[4,5]cyclopenta[1,2-c]pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kang, H.J, Diberto, J.F, Kapolka, N.J, Gumpper, R.H, Olsen, R.H.J, Huang, X.P, Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-10-30 | | Release date: | 2025-02-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure-guided design of a peripherally restricted chemogenetic system.
Cell, 187, 2024
|
|
8UUJ
 
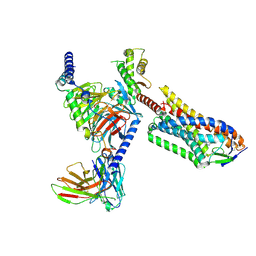 | | CryoEM Structure of HCA2 DREADD Gi1 in complex with FCH-2296413 | | Descriptor: | (3M,4aR,5aR)-3-(1H-tetrazol-5-yl)-4,4a,5,5a-tetrahydro-1H-cyclopropa[4,5]cyclopenta[1,2-c]pyrazole, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kang, H.J, Diberto, J.F, Kapolka, N.J, Gumpper, R.H, Olsen, R.H.J, Huang, X.P, Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-11-01 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided design of a peripherally restricted chemogenetic system.
Cell, 187, 2024
|
|
8U02
 
 | | CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant and dopamine | | Descriptor: | D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kapolka, N.J, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-08-28 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state.
Nat Commun, 15, 2024
|
|
8TZQ
 
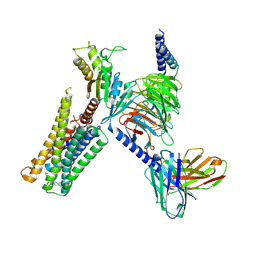 | | CryoEM structure of D2 dopamine receptor in complex with GoA KE Mutant, scFv16, and dopamine | | Descriptor: | D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kapolka, N.J, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-08-27 | | Release date: | 2024-08-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state.
Nat Commun, 15, 2024
|
|
8FN0
 
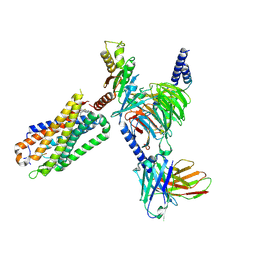 | | CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, DiBerto, J.F, Olsen, R.H.J, Kang, H, Slocum, S.T, Zhang, S, Strachan, R.T, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-12-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Neurotensin Receptor Allosterism Revealed in Complex with a Biased Allosteric Modulator.
Biochemistry, 62, 2023
|
|
8FMZ
 
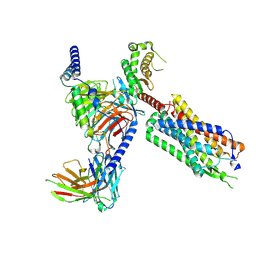 | | Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, MiniGq, ... | | Authors: | Krumm, B.E, Diberto, J.F, Olsen, R.H.J, Kang, H, Slocum, S.T, Zhang, S, Strachan, R.T, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-12-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Neurotensin Receptor Allosterism Revealed in Complex with a Biased Allosteric Modulator.
Biochemistry, 62, 2023
|
|
8FN1
 
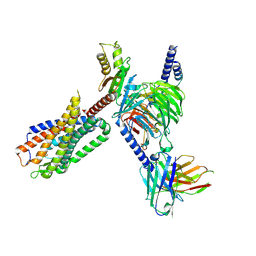 | | CryoEM structure of Go-coupled NTSR1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Krumm, B.E, DiBerto, J.F, Olsen, R.H.J, Kang, H, Slocum, S.T, Zhang, S, Strachan, R.T, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-12-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Neurotensin Receptor Allosterism Revealed in Complex with a Biased Allosteric Modulator.
Biochemistry, 62, 2023
|
|
6B73
 
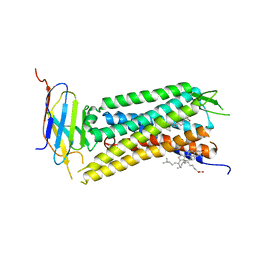 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
2I6V
 
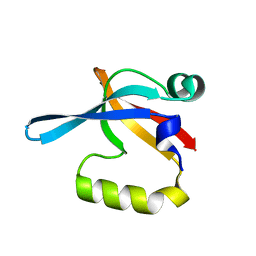 | | PDZ domain of EpsC from Vibrio cholerae, residues 219-305 | | Descriptor: | General secretion pathway protein C | | Authors: | Korotkov, K.V, Krumm, B, Bagdasarian, M, Hol, W.G.J. | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and Functional Studies of EpsC, a Crucial Component of the Type 2 Secretion System from Vibrio cholerae.
J.Mol.Biol., 363, 2006
|
|
1P9W
 
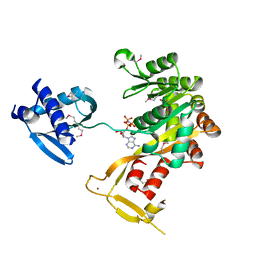 | | Crystal Structure of Vibrio cholerae putative NTPase EpsE | | Descriptor: | General secretion pathway protein E, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ZINC ION | | Authors: | Robien, M.A, Krumm, B.E, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2003-05-12 | | Release date: | 2003-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae
J.Mol.Biol., 333, 2003
|
|
1P9R
 
 | | Crystal Structure of Vibrio cholerae putative NTPase EpsE | | Descriptor: | CHLORIDE ION, General secretion pathway protein E, ZINC ION | | Authors: | Robien, M.A, Krumm, B.E, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2003-05-12 | | Release date: | 2003-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae
J.Mol.Biol., 333, 2003
|
|
2I4S
 
 | | PDZ domain of EpsC from Vibrio cholerae, residues 204-305 | | Descriptor: | General secretion pathway protein C | | Authors: | Korotkov, K.V, Krumm, B, Bagdasarian, M, Hol, W.G.J. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Functional Studies of EpsC, a Crucial Component of the Type 2 Secretion System from Vibrio cholerae.
J.Mol.Biol., 363, 2006
|
|
7CMU
 
 | | Dopamine Receptor D3R-Gi-Pramipexole complex | | Descriptor: | (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
