2VPP
 
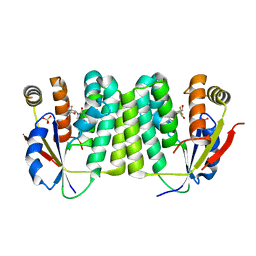 | | Drosophila melanogaster deoxyribonucleoside kinase successfully activates gemcitabine in transduced cancer cell lines | | Descriptor: | DEOXYNUCLEOSIDE KINASE, GEMCITABINE, SULFATE ION | | Authors: | Knecht, W, Mikkelsen, N.E, Clausen, A, Willer, M, Gojkovic, Z, Piskur, J. | | Deposit date: | 2008-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Drosophila Melanogaster Deoxyribonucleoside Kinase Activates Gemcitabine.
Biochem.Biophys.Res.Commun., 382, 2009
|
|
6ZCT
 
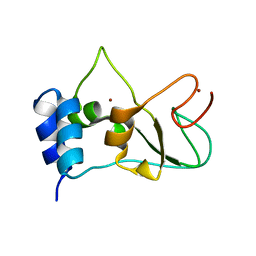 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
2OCP
 
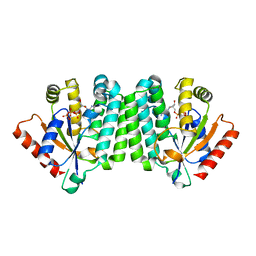 | | Crystal Structure of Human Deoxyguanosine Kinase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxyguanosine kinase | | Authors: | Johansson, K, Ramaswamy, S, Ljungkrantz, C, Knecht, W, Piskur, J, Munch-Petersen, B, Eriksson, S, Eklund, H. | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Substrate Specificities of Cellular Deoxyribonucleoside Kinases.
Nat.Struct.Biol., 8, 2001
|
|
1J90
 
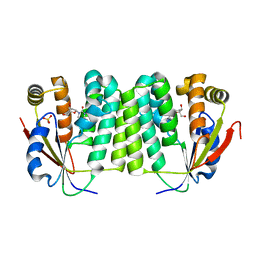 | | Crystal Structure of Drosophila Deoxyribonucleoside Kinase | | Descriptor: | 2'-DEOXYCYTIDINE, Deoxyribonucleoside kinase, SULFATE ION | | Authors: | Johansson, K, Ramaswamy, S, Ljungkrantz, C, Knecht, W, Piskur, J, Munch-Petersen, B, Eriksson, S, Eklund, H. | | Deposit date: | 2001-05-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for substrate specificities of cellular deoxyribonucleoside kinases.
Nat.Struct.Biol., 8, 2001
|
|
1OT3
 
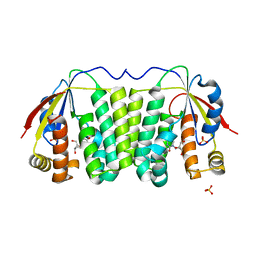 | | Crystal structure of Drosophila deoxyribonucleotide kinase complexed with the substrate deoxythymidine | | Descriptor: | Deoxyribonucleoside Kinase, SULFATE ION, THYMIDINE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-21 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway: Studies of the Drosophila Deoxyribonucleoside Kinase.
Biochemistry, 42, 2003
|
|
1OE0
 
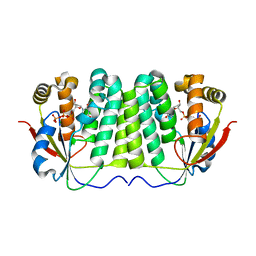 | | CRYSTAL STRUCTURE OF DROSOPHILA DEOXYRIBONUCLEOSIDE KINASE IN COMPLEX WITH DTTP | | Descriptor: | DEOXYRIBONUCLEOSIDE KINASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-17 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway:Studies of the Drosophila Deoxyribonucleoside Kinase
Biochemistry, 42, 2003
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
1ZMX
 
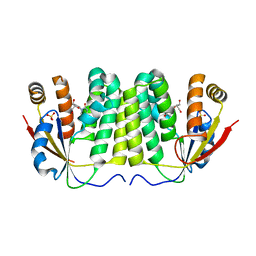 | | Crystal structure of D. melanogaster deoxyribonucleoside kinase N64D mutant in complex with thymidine | | Descriptor: | Deoxynucleoside kinase, SULFATE ION, THYMIDINE | | Authors: | Welin, M, Skovgaard, T, Knecht, W, Berenstein, D, Munch-Petersen, B, Piskur, J, Eklund, H. | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the changed substrate specificity of Drosophila melanogaster deoxyribonucleoside kinase mutant N64D.
Febs J., 272, 2005
|
|
1ZM7
 
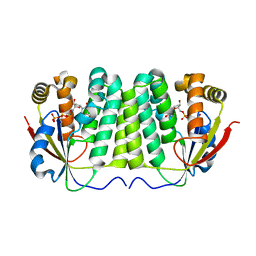 | | Crystal structure of D. melanogaster deoxyribonucleoside kinase mutant N64D in complex with dTTP | | Descriptor: | Deoxynucleoside kinase, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Welin, M, Skovgaard, T, Knecht, W, Berenstein, D, Munch-Petersen, B, Piskur, J, Eklund, H. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the changed substrate specificity of Drosophila melanogaster deoxyribonucleoside kinase mutant N64D.
Febs J., 272, 2005
|
|
4BVV
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, APOLIPOPROTEIN(A), SULFATE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVW
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1,2,3,4-tetrahydroisoquinoline-6-carboxylic acid, 1,2-ETHANEDIOL, APOLIPOPROTEIN(A), ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVC
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-PIPERIDYL)-N-[2-(TRIFLUOROMETHOXY)PHENYL]SULFONYL-PROPANAMIDE, APOLIPOPROTEIN(A), CHLORIDE ION, ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4CR5
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-chloroquinolin-2(1H)-one, COAGULATION FACTOR XIA, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-25 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CRC
 
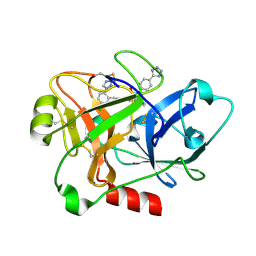 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | (2S)-2-[[(E)-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-3-phenyl-N-[4-(1H-1,2,3,4-tetrazol-5-yl)phenyl]propanamide, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4AEL
 
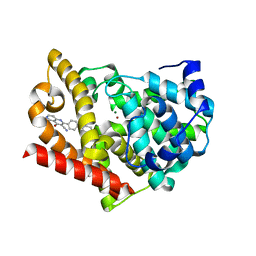 | | PDE10A in complex with the inhibitor AZ5 | | Descriptor: | 2-(2'-ETHOXYBIPHENYL-4-YL)-4-HYDROXY-1,6-NAPHTHYRIDINE-3-CARBONITRILE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Bauer, U, Giordanetto, F, Bauer, M, OMahony, G, Johansson, K.E, Knecht, W, Hartleib-Geschwindner, J, Toppner Carlsson, E, Enroth, C, Sjogren, T. | | Deposit date: | 2012-01-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of 4-Hydroxy-1,6-Naphthyridine-3-Carbonitrile Derivatives as Novel Pde10A Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4BV7
 
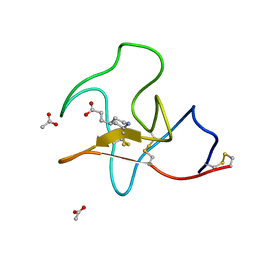 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVD
 
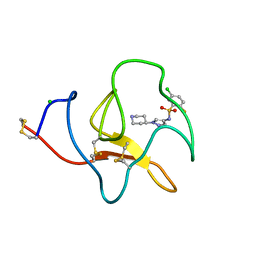 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 5-chloro-2-fluoro-N-[1-(4-piperidyl)pyrazol-4-yl]benzenesulfonamide, APOLIPOPROTEIN(A), CHLORIDE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BV5
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 4-(aminomethyl)-N-(benzenesulfonyl)cyclohexanecarboxamide, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4CRE
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-chloro-4-methyl-1H-quinolin-2-one, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CRG
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-carbamimidoyl-N-phenyl-4-(pyrimidin-2-ylamino)naphthalene-2-carboxamide, COAGULATION FACTOR XI, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CR9
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 4-methylquinoline-2,6-diamine, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CRA
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | COAGULATION FACTOR XI, GLYCEROL, N-[(1S)-2-[(2-amino-5-quinolyl)methylamino]-1-benzyl-2-oxo-ethyl]-4-hydroxy-2-oxo-1H-quinoline-6-carboxamide, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
