1KD8
 
 | | X-RAY STRUCTURE OF THE COILED COIL GCN4 ACID BASE HETERODIMER ACID-d12Ia16V BASE-d12La16L | | Descriptor: | GCN4 ACID BASE HETERODIMER ACID-d12Ia16V, GCN4 ACID BASE HETERODIMER BASE-d12La16L | | Authors: | Keating, A.E, Malashkevich, V.N, Tidor, B, Kim, P.S. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KD9
 
 | | X-RAY STRUCTURE OF THE COILED COIL GCN4 ACID BASE HETERODIMER ACID-d12La16L BASE-d12La16L | | Descriptor: | GCN4 ACID BASE HETERODIMER ACID-d12La16L, GCN4 ACID BASE HETERODIMER BASE-d12La16L | | Authors: | Keating, A.E, Malashkevich, V.N, Tidor, B, Kim, P.S. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KDD
 
 | | X-ray structure of the coiled coil GCN4 ACID BASE HETERODIMER ACID-d12La16I BASE-d12La16L | | Descriptor: | GCN4 ACID BASE HETERODIMER ACID-d12La16I, GCN4 ACID BASE HETERODIMER BASE-d12La16L | | Authors: | Keating, A.E, Malashkevich, V.N, Tidor, B, Kim, P.S. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7LXE
 
 | |
1XOF
 
 | | Heterooligomeric Beta Beta Alpha Miniprotein | | Descriptor: | BBAhetT1 | | Authors: | Ali, M.H, Taylor, C.M, Grigoryan, G, Allen, K.N, Imperiali, B, Keating, A.E. | | Deposit date: | 2004-10-06 | | Release date: | 2005-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of a Heterospecific, Tetrameric, 21-Residue Miniprotein with Mixed alpha/beta Structure.
Structure, 13, 2005
|
|
1L5A
 
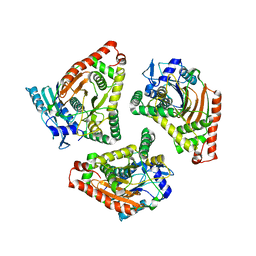 | | Crystal Structure of VibH, an NRPS Condensation Enzyme | | Descriptor: | amide synthase | | Authors: | Keating, T.A, Marshall, C.G, Walsh, C.T, Keating, A.E. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains.
Nat.Struct.Biol., 9, 2002
|
|
8CZH
 
 | | Human BAK in complex with the dM2 peptide | | Descriptor: | Bcl-2 homologous antagonist/killer, DM2 peptide | | Authors: | Aguilar, F, Keating, A.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization.
Structure, 31, 2023
|
|
8CZF
 
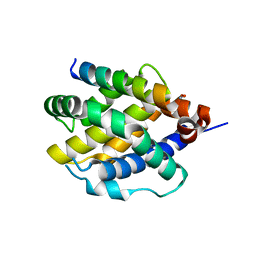 | | Human BAK in complex with the dF2 peptide | | Descriptor: | Bcl-2 homologous antagonist/killer, DF2 peptide | | Authors: | Aguilar, F, Keating, A.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization.
Structure, 31, 2023
|
|
8CZG
 
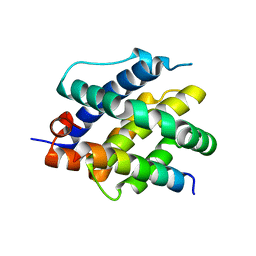 | | Human BAK in complex with the dF3 peptide | | Descriptor: | Bcl-2 homologous antagonist/killer, dF3 peptide | | Authors: | Aguilar, F, Keating, A.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization.
Structure, 31, 2023
|
|
6E3J
 
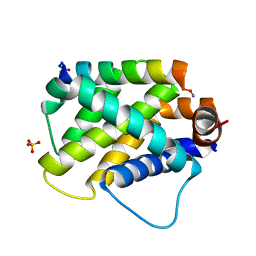 | |
6E3I
 
 | |
6UAB
 
 | |
7LXF
 
 | |
6UA3
 
 | |
6MBB
 
 | | Human Bfl-1 in complex with the designed peptide dF1 | | Descriptor: | Bcl-2-related protein A1, dF1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBE
 
 | | Human Mcl-1 in complex with the designed peptide dM7 | | Descriptor: | CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, dM7 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBC
 
 | | Human Bfl-1 in complex with the designed peptide dF4 | | Descriptor: | Bcl-2-related protein A1, dF4 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBD
 
 | | Human Mcl-1 in complex with the designed peptide dM1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, dM1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
2PQK
 
 | | X-ray crystal structure of human Mcl-1 in complex with Bim BH3 | | Descriptor: | Bim BH3 peptide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Bare, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3HE5
 
 | |
3HE4
 
 | |
3KJ1
 
 | | Mcl-1 in complex with Bim BH3 mutant I2dA | | Descriptor: | ACETATE ION, Bcl-2-like protein 11, CHLORIDE ION, ... | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KZ0
 
 | | MCL-1 complex with MCL-1-specific selected peptide | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 specific peptide MB7, SULFATE ION, ... | | Authors: | Dutta, S, Fire, E, Grant, R.A, Sauer, R.T, Keating, A.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL.
J.Mol.Biol., 398, 2010
|
|
3KJ2
 
 | | Mcl-1 in complex with Bim BH3 mutant F4aE | | Descriptor: | ACETATE ION, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KJ0
 
 | | Mcl-1 in complex with Bim BH3 mutant I2dY | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
