2Z9U
 
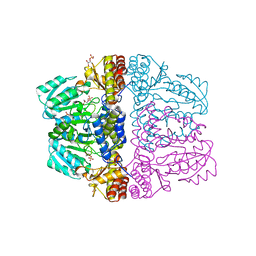 | | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti at 2.0 A resolution | | Descriptor: | Aspartate aminotransferase, GLYCEROL, SULFATE ION | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2NTM
 
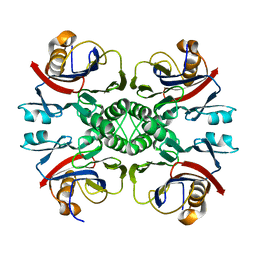 | |
2Z9V
 
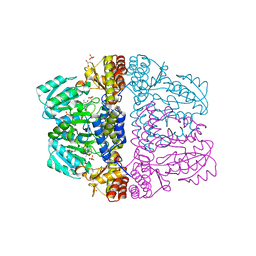 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2NTL
 
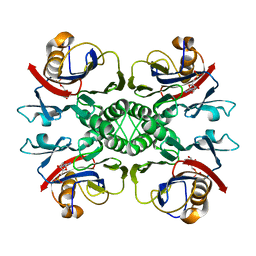 | | Crystal structure of PurO/AICAR from Methanothermobacter thermoautotrophicus | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, IMP cyclohydrolase | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2NTK
 
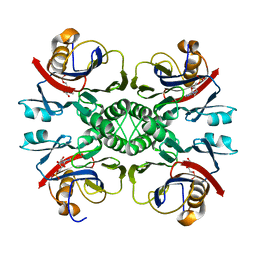 | | Crystal structure of PurO/IMP from Methanothermobacter thermoautotrophicus | | Descriptor: | IMP cyclohydrolase, INOSINIC ACID | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
1V3H
 
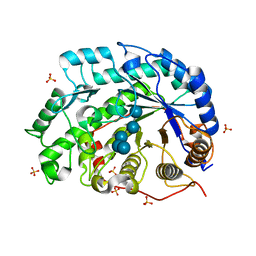 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
1V3I
 
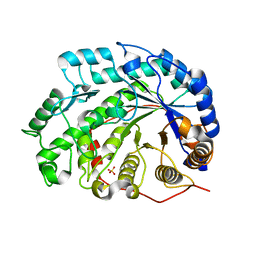 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
4Z3S
 
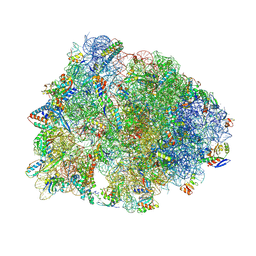 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic A201A, mRNA and three tRNAs in the A, P and E sites at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 3'-deoxy-3'-{[(2E)-3-(4-{[(4Z)-6-O-(6-deoxy-3,4-di-O-methyl-alpha-D-mannopyranosyl)-5-O-methyl-alpha-D-threo-hex-4-enofuranosyl]oxy}phenyl)-2-methylprop-2-enoyl]amino}-N,N-dimethyladenosine, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-03-31 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
7KNW
 
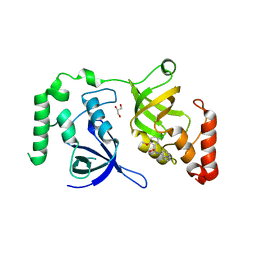 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNX
 
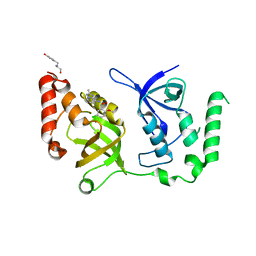 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
8FE9
 
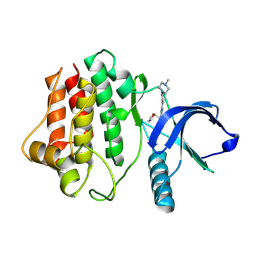 | | Crystal structure of Ack1 kinase K161Q mutant in complex with the selective inhibitor (R)-9b | | Descriptor: | Activated CDC42 kinase 1, N-[(1S)-1-benzyl-2-[(6-chloro-2-oxo-1H-quinolin-4-yl)methylamino]-2-oxo-ethyl]-4-hydroxy-2-oxo-1H-quinoline-6-carbo | | Authors: | Paung, Y, Kan, Y, Seeliger, M.S, Miller, W.T. | | Deposit date: | 2022-12-06 | | Release date: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical Studies of Systemic Lupus Erythematosus-Associated Mutations in Nonreceptor Tyrosine Kinases Ack1 and Brk.
Biochemistry, 62, 2023
|
|
5W0P
 
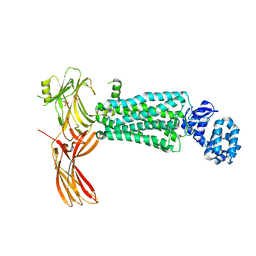 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
7RQ8
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
7MD7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with triphenylphosphonium analog of chloramphenicol CAM-C4-TPP and protein Y (YfiA) at 2.80A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Pavlova, J.A, Lukianov, D.A, Tereshchenkov, A.G, Makarov, G.I, Khairullina, Z.Z, Tashlitsky, V.N, Paleskava, A, Konevega, A.L, Bogdanov, A.A, Osterman, I.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2021-04-03 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Triphenylphosphonium Analog of Chloramphenicol upon the Bacterial Ribosome.
Antibiotics, 10, 2021
|
|
7RQ9
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
8T8B
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site formyl-MAI-tripeptidyl-tRNA analog ACCA-IAMf at 2.65A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Thaler, J, Syroegin, E.A, Breuker, K, Polikanov, Y.S, Micura, R. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Practical Synthesis of N -Formylmethionylated Peptidyl-tRNA Mimics.
Acs Chem.Biol., 18, 2023
|
|
8T8C
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site formyl-MFI-tripeptidyl-tRNA analog ACCA-IFMf at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Thaler, J, Syroegin, E.A, Breuker, K, Polikanov, Y.S, Micura, R. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Practical Synthesis of N -Formylmethionylated Peptidyl-tRNA Mimics.
Acs Chem.Biol., 18, 2023
|
|
5DJ5
 
 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
4V9S
 
 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
1VFU
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 amylase 2/gamma-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
5KCS
 
 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Evernimycin, mRNA, TetM and P-site tRNA at 3.9A resolution | | Descriptor: | (2R,3R,4R,6S)-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7S,7aR)-6-{[(2S,3R,4R,5S,6R)-2-{[(2R,3S,4S,5S,6S)-6-({(2R,3aS,3a'R,6S,7R,7'R,7aS,7a'S)-7'-[(2,4-dihydroxy-6-methylbenzoyl)oxy]-7-hydroxyoctahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-4-hydroxy-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-3-hydroxy-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4',7-dihydroxy-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-4-{[(2R,4S,5R,6S)-5-methoxy-4,6-dimethyl-4-nitrotetrahydro-2H-pyran-2-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KCR
 
 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Avilamycin C, mRNA and P-site tRNA at 3.6A resolution | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V9R
 
 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
