2DKJ
 
 | | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, serine hydroxymethyltransferase | | Authors: | Kai, K, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase
To be Published
|
|
7E7F
 
 | | Human CYP11B1 mutant in complex with metyrapone | | Descriptor: | CHOLIC ACID, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Mukai, K, Sugimoto, H, Reiko, S, Matsuura, T, Hishiki, T, Kagawa, N. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Spatially restricted substrate-binding site of cortisol-synthesizing CYP11B1 limits multiple hydroxylations and hinders aldosterone synthesis.
Curr Res Struct Biol, 3, 2021
|
|
4K0W
 
 | | X-ray crystal structure of OXA-23 A220 duplication clinical variant | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Beta-lactamase, ... | | Authors: | Kaitany, K.J, Klinger, N.V, Leonard, D.A, Powers, R.A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the Class D Carbapenemases OXA-23 and OXA-146: Mechanistic Basis of Activity against Carbapenems, Extended-Spectrum Cephalosporins, and Aztreonam.
Antimicrob.Agents Chemother., 57, 2013
|
|
4X6X
 
 | | Human soluble epoxide hydrolase in complex with a three substituted cyclopropane derivative | | Descriptor: | 3-{4-[(1-{[(1s,2R,3S)-2,3-diphenylcyclopropyl]carbamoyl}piperidin-4-yl)oxy]phenyl}propanoic acid, Bifunctional epoxide hydrolase 2 | | Authors: | Chiyo, N, Takai, K, Ishii, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional rational approach to the discovery of potent substituted cyclopropyl urea soluble epoxide hydrolase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2VQE
 
 | | Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Kurata, S, Weixlbaumer, A, Ohtsuki, T, Shimazaki, T, Wada, T, Kirino, Y, Takai, K, Watanabe, K, Ramakrishnan, V, Suzuki, T. | | Deposit date: | 2008-03-13 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modified Uridines with C5-Methylene Substituents at the First Position of the tRNA Anticodon Stabilize U.G Wobble Pairing During Decoding.
J.Biol.Chem., 283, 2008
|
|
2VQF
 
 | | Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Kurata, S, Weixlbaumer, A, Ohtsuki, T, Shimazaki, T, Wada, T, Kirino, Y, Takai, K, Watanabe, K, Ramakrishnan, V, Suzuki, T. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modified Uridines with C5-Methylene Substituents at the First Position of the tRNA Anticodon Stabilize U.G Wobble Pairing During Decoding.
J.Biol.Chem., 283, 2008
|
|
5GUA
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138Y mutant | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU9
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138I mutant | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU8
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) wild type | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, SODIUM ION | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7DCZ
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-4H-1,3-thiazin-4-yl]-4- fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Koriyama, Y, Hori, A, Ito, H, Yonezawa, S, Baba, Y, Tanimoto, N, Ueno, T, Yamamoto, S, Yamamoto, T, Asada, N, Morimoto, K, Einaru, S, Sakai, K, Kanazu, T, Matsuda, A, Yamaguchi, Y, Oguma, T, Timmers, M, Tritsmans, L, Kusakabe, K.I, Kato, A, Sakaguchi, G. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Atabecestat (JNJ-54861911): A Thiazine-Based beta-Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor Advanced to the Phase 2b/3 EARLY Clinical Trial.
J.Med.Chem., 64, 2021
|
|
7WAG
 
 | | Crystal structure of MurJ squeezed form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAX
 
 | | MurJ inward occluded form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4S)-2-METHYL-2,4-PENTANEDIOL, lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAW
 
 | | MurJ inward closed form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
6LEO
 
 | | Crystal structure of thiosulfate transporter YeeE from Spirochaeta thermophila | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Takeuchi, A, Uchino, S, Sugano, Y. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
1A2B
 
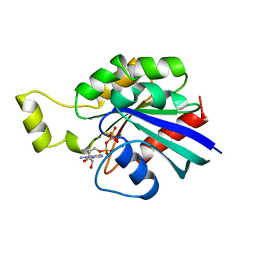 | | HUMAN RHOA COMPLEXED WITH GTP ANALOGUE | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA | | Authors: | Ihara, K, Muraguchi, S, Kato, M, Shimizu, T, Shirakawa, M, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1997-12-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue.
J.Biol.Chem., 273, 1998
|
|
2H9V
 
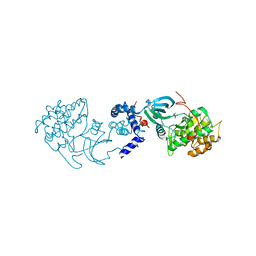 | | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Miwa, Y, Kasa, M, Kitano, K, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2006-06-12 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632
J.Biochem.(Tokyo), 140, 2006
|
|
1CXZ
 
 | | CRYSTAL STRUCTURE OF HUMAN RHOA COMPLEXED WITH THE EFFECTOR DOMAIN OF THE PROTEIN KINASE PKN/PRK1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PROTEIN (HIS-TAGGED TRANSFORMING PROTEIN RHOA(0-181)), ... | | Authors: | Maesaki, R, Ihara, K, Shimizu, T, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-08-31 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1.
Mol.Cell, 4, 1999
|
|
1UIX
 
 | | Coiled-coil structure of the RhoA-binding domain in Rho-kinase | | Descriptor: | Rho-associated kinase | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel coiled-coil association of the RhoA-binding domain in Rho-kinase
J.Biol.Chem., 278, 2003
|
|
5YHF
 
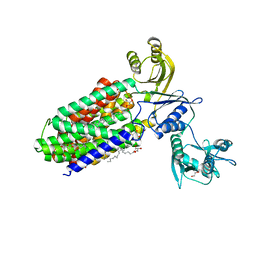 | | Crystal structure of SecDF in Super-membrane-facing form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Protein translocase subunit SecDF | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Furukawa, A. | | Deposit date: | 2017-09-28 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Remote Coupled Drastic beta-Barrel to beta-Sheet Transition of the Protein Translocation Motor.
Structure, 26, 2018
|
|
6LEP
 
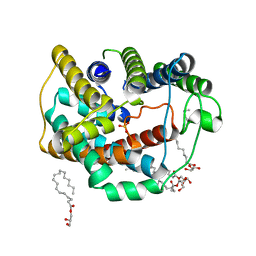 | | Crystal structure of thiosulfate transporter YeeE inactive mutant - C91A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Sugano, Y, Takeuchi, A, Uchino, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
5YVQ
 
 | | Complex of Mu phage tail fiber and its chaperone | | Descriptor: | GLYCEROL, Tail fiber assembly protein U, Tail fiber protein S | | Authors: | Takeda, S, Sakai, K, Iwazaki, T, Yamashita, E, Nakagawa, A. | | Deposit date: | 2017-11-27 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres.
Nat Microbiol, 4, 2019
|
|
1DPF
 
 | | CRYSTAL STRUCTURE OF A MG-FREE FORM OF RHOA COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, RHOA | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-12-27 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An open conformation of switch I revealed by the crystal structure of a Mg2+-free form of RHOA complexed with GDP. Implications for the GDP/GTP exchange mechanism.
J.Biol.Chem., 275, 2000
|
|
4X6Y
 
 | |
3A8Q
 
 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
