8B57
 
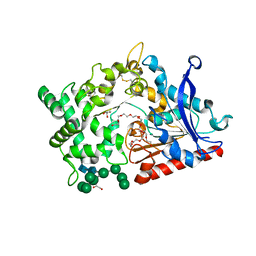 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-09-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
9BOL
 
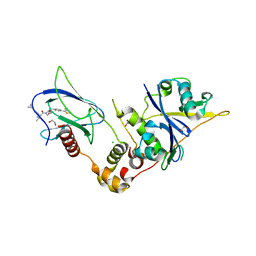 | | Crystal structure of the complex between VHL, ElonginB, ElonginC, and compound 5 | | Descriptor: | (4R)-1-[(2S)-2-(4-cyclopropyl-1H-1,2,3-triazol-1-yl)-3,3-dimethylbutanoyl]-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Murray, J.M, Wu, H, Fuhrmann, J, Fairbrother, W.J, DiPasquale, A. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potency-enhanced peptidomimetic VHL ligands with improved oral bioavailability
To Be Published
|
|
8BEI
 
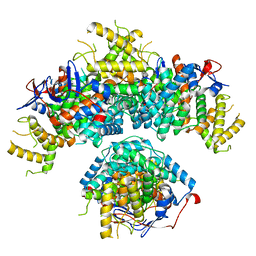 | |
5D6Y
 
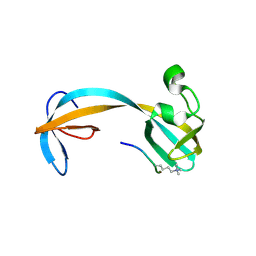 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
6OQI
 
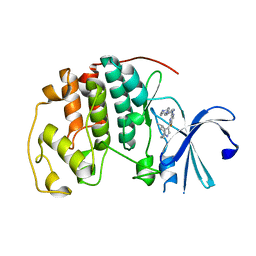 | | CDK2 in complex with Cpd14 (5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine) | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4S)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5D9A
 
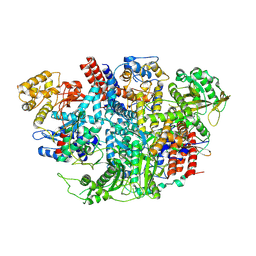 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P212121 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
8CGX
 
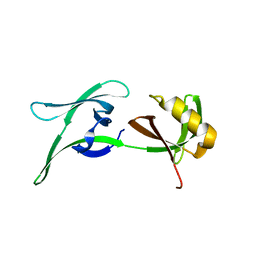 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using XDS | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CGY
 
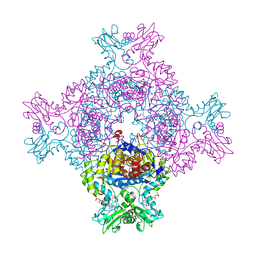 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; XDS processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD5
 
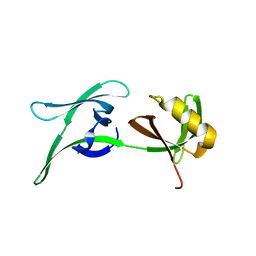 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C9J
 
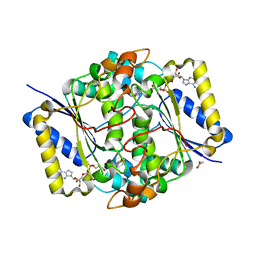 | | Crystal structure of human NQO1 by serial femtosecond crystallography | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Pey, A, Botha, S, Ros, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
5ERY
 
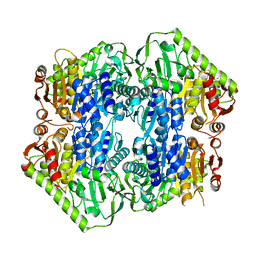 | | Crystal Structure of APO MenD from M. tuberculosis - P212121 | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Johnston, J.M, Jirgis, E.N.M, Bashiri, G, Bulloch, E.M.M, Baker, E.N. | | Deposit date: | 2015-11-16 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Views along the Mycobacterium tuberculosis MenD Reaction Pathway Illuminate Key Aspects of Thiamin Diphosphate-Dependent Enzyme Mechanisms.
Structure, 24, 2016
|
|
5EIQ
 
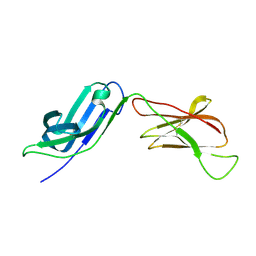 | |
5ES8
 
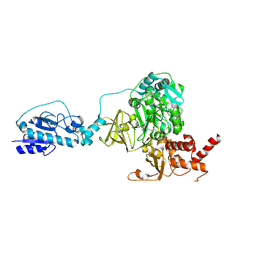 | | Crystal structure of the initiation module of LgrA in the thiolation state | | Descriptor: | Linear gramicidin synthetase subunit A, [(3~{R})-4-[[3-[2-[[(2~{S})-2-azanyl-3-methyl-butanoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Reimer, J.M, Aloise, M.N, Schmeing, T.M. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase.
Nature, 529, 2016
|
|
5ES5
 
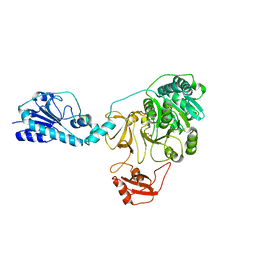 | |
7S81
 
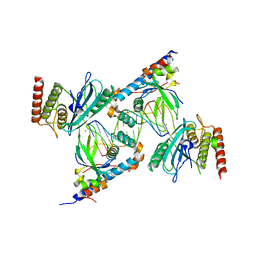 | | Structure of human PARP1 domains (Zn1, Zn3, WGR, HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*CP*GP*CP*AP*T)-3'), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S6H
 
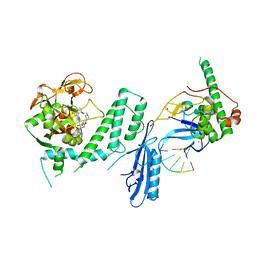 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
1KUJ
 
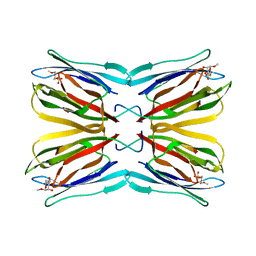 | | Crystal structure of Jacalin complexed with 1-O-methyl-alpha-D-mannose | | Descriptor: | JACALIN ALPHA CHAIN, JACALIN BETA CHAIN, methyl alpha-D-mannopyranoside | | Authors: | Bourne, Y, Astoul, C.H, Zamboni, V, Peumans, W.J, Menu-Bouaouiche, L, Van Damme, E.J.M, Barre, A, Rouge, P. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose.
Biochem.J., 364, 2002
|
|
7S6M
 
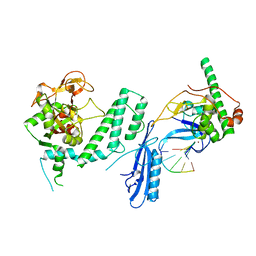 | |
7S68
 
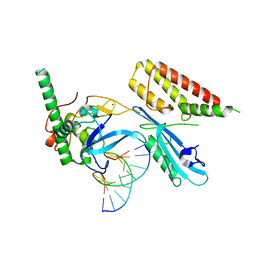 | | Structure of human PARP1 domains (Zn1, Zn3, WGR and HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3'), Fusion of PARP1 zinc fingers 1 and 3 (Zn1, Zn3), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
5ES6
 
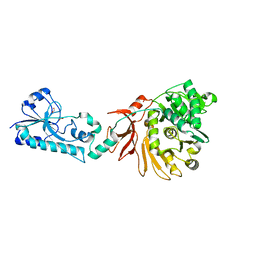 | |
5ES7
 
 | | Crystal structure of the F-A domains of the LgrA initiation module soaked with FON, AMPcPP, and valine. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthetase subunit A, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Reimer, J.M, Aloise, M.N, Schmeing, T.M. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase.
Nature, 529, 2016
|
|
7RZI
 
 | | Insulin Degrading Enzyme pC/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZH
 
 | | Insulin Degrading Enzyme O/O | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZE
 
 | | Insulin Degrading Enzyme pO/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZF
 
 | | Insulin Degrading Enzyme O/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
