4QX2
 
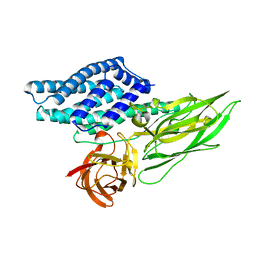 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZD1
 
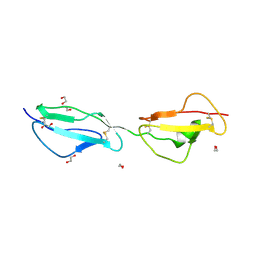 | | STRUCTURE OF THE TWO C-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 2 | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 2 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6MJ5
 
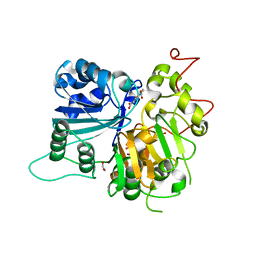 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ519 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-nitroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6N0O
 
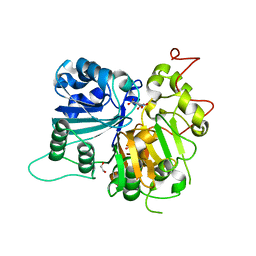 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ523 | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
6MWW
 
 | | LasR LBD:BB0126 complex | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1R,3R,5R)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6N19
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ578 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-carboxybutanoyl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6MYZ
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ520 | | Descriptor: | 1,2-ETHANEDIOL, 4-oxo-8-phenyl-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Structure of Tdp1 catalytic domain in complex with compound XZ520
To Be Published
|
|
6MVN
 
 | | LasR LBD L130F:3OC10HSL complex | | Descriptor: | 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants driving homoserine lactone ligand selection in thePseudomonas aeruginosaLasR quorum-sensing receptor.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6MWL
 
 | | LasR LBD:mBTL complex | | Descriptor: | 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-29 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MWZ
 
 | | LasR LBD T75V/Y93F/A127W:BB0126 | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1S,3S,5S)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, ALA-HIS-HIS-HIS-HIS-ALA, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MVM
 
 | | LasR LBD L130F:3OC14HSL complex | | Descriptor: | 3-oxo-N-[(3S)-2-oxooxolan-3-yl]tetradecanamide, Transcriptional regulator LasR | | Authors: | Paczkowski, J.E, Bassler, B.L. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MZ0
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ530 | | Descriptor: | 1,2-ETHANEDIOL, 5-hydroxypyrazine-2,3-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structure of Tdp1 catalytic domain in complex with compound XZ530
To Be Published
|
|
6N0R
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ572 | | Descriptor: | 1,2-ETHANEDIOL, 4-(methylamino)benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
6N17
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ577 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-carboxypropanoyl)amino]benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6MWH
 
 | | LasR LBD:BB0020 complex | | Descriptor: | 2-(3-bromophenoxy)-N-[(1S,2S,3R,5S)-2-hydroxybicyclo[3.1.0]hexan-3-yl]acetamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-29 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6N0D
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ575 | | Descriptor: | 1,2-ETHANEDIOL, 4-fluorobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain in complex with compound XZ575
To Be Published
|
|
6NSV
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | Descriptor: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2019-01-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
6NEF
 
 | | Outer Membrane Cytochrome S Filament from Geobacter Sulfurreducens | | Descriptor: | C-type cytochrome OmcS, HEME C, MAGNESIUM ION | | Authors: | Filman, D.J, Marino, S.F, Ward, J.E, Yang, L, Mester, Z, Bullitt, E, Lovley, D.R, Strauss, M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire.
Commun Biol, 2, 2019
|
|
6NIZ
 
 | |
6NPN
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine-3-hydroxyindazole inhibitor | | Descriptor: | (7R)-7-methyl-2-[(3-oxo-2,3-dihydro-1H-indazol-6-yl)amino]-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, dos Reis, C.V, Takarada, J.E, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine-3-hydroxyindazole inhibitor
To Be Published
|
|
6N0N
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ574 | | Descriptor: | 1,2-ETHANEDIOL, 4-methylbenzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
6OFE
 
 | |
6OFG
 
 | | In vitro polymerized PrgI V67A filaments | | Descriptor: | Protein PrgI | | Authors: | Guo, E.Z, Galan, J.E. | | Deposit date: | 2019-03-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A polymorphic helix of a Salmonella needle protein relays signals defining distinct steps in type III secretion.
Plos Biol., 17, 2019
|
|
6OL8
 
 | | Crystal structure of NDM-12 metallo-beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-12, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
6OFH
 
 | |
