8JAX
 
 | |
7OZU
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with A | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6TRU
 
 | |
5G2N
 
 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
7ZVF
 
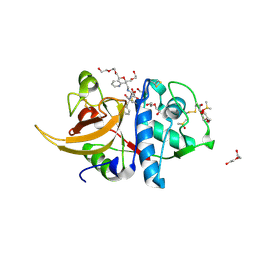 | | Crystal structure of human cathepsin L in complex with covalently bound CLIK148 | | Descriptor: | (2S)-N-[(2S)-1-(dimethylamino)-1-oxidanylidene-3-phenyl-propan-2-yl]-2-oxidanyl-N'-(2-pyridin-2-ylethyl)butanediamide, 1,2-ETHANEDIOL, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Tsuge, H, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
6Q42
 
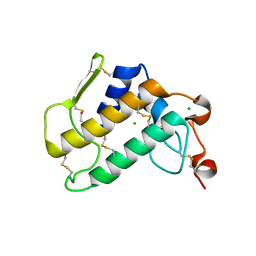 | | Crystal Structure of Human Pancreatic Phospholipase A2 | | Descriptor: | CHLORIDE ION, Phospholipase A2 | | Authors: | Saul, F, Haouz, A, Lambeau, G, Theze, J. | | Deposit date: | 2018-12-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PLA2G1B is involved in CD4 anergy and CD4 lymphopenia in HIV-infected patients.
J.Clin.Invest., 130, 2020
|
|
6TQT
 
 | | The crystal structure of the MSP domain of human MOSPD2. | | Descriptor: | 1,2-ETHANEDIOL, Motile sperm domain-containing protein 2, PHOSPHATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6DGT
 
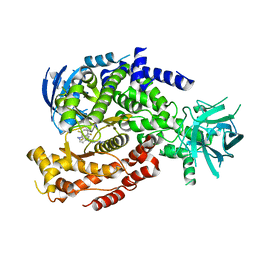 | | Selective PI3K beta inhibitor bound to PI3K delta | | Descriptor: | 4-[1-(5,8-difluoroquinolin-4-yl)-2-methyl-4-(4H-1,2,4-triazol-3-yl)-1H-benzimidazol-6-yl]-3-fluoropyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J, Villasenor, A, McGrath, M. | | Deposit date: | 2018-05-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Atropisomerism by Design: Discovery of a Selective and Stable Phosphoinositide 3-Kinase (PI3K) beta Inhibitor.
J. Med. Chem., 61, 2018
|
|
6DFR
 
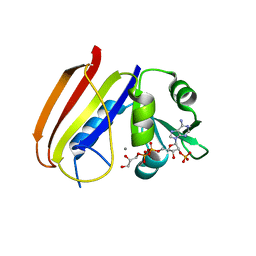 | |
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6TY3
 
 | | FAK structure from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.32 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
6H35
 
 | | Myxococcus xanthus MglA bound to GDP and Pi with mixed inactive and active switch region conformations | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Mutual gliding-motility protein MglA, ... | | Authors: | Varela, P.F, Galicia, C, Cherfils, J. | | Deposit date: | 2018-07-17 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MglA functions as a three-state GTPase to control movement reversals of Myxococcus xanthus.
Nat Commun, 10, 2019
|
|
7U8C
 
 | | Crystal structure of Mesothelin C-terminal peptide-MORAb 15B6 FAB complex | | Descriptor: | MORab 15B6 Fab heavy chain, MORab 15B6 Fab light chain, Mesothelin, ... | | Authors: | Zhan, J, Esser, L, Xia, D. | | Deposit date: | 2022-03-08 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Highly active CAR T cells that bind to a juxtamembrane region of mesothelin and are not blocked by shed mesothelin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6GXB
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor JS13 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, ~{N}-[4-[4-[(4-sulfamoylphenyl)carbamoylamino]phenoxy]butyl]ethanamide | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibitor-Polymer Conjugates as a Versatile Tool for Detection and Visualization of Cancer-Associated Carbonic Anhydrase Isoforms
Acs Omega, 4, 2019
|
|
5HI8
 
 | | Structure of T-type Phycobiliprotein Lyase CpeT from Prochlorococcus phage P-HM1 | | Descriptor: | ACETATE ION, Antenna protein, MAGNESIUM ION | | Authors: | Gasper, R, Schwach, J, Frankenberg-Dinkel, N, Hofmann, E. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct Features of Cyanophage-encoded T-type Phycobiliprotein Lyase Phi CpeT: THE ROLE OF AUXILIARY METABOLIC GENES.
J. Biol. Chem., 292, 2017
|
|
6PUD
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-5'OH-Pentyl-5-OP-U | | Descriptor: | 6-[(5-hydroxypentyl)amino]-5-[(E)-propylideneamino]pyrimidine-2,4(1H,3H)-dione, Beta-2-microglobulin, Human TCR alpha chain, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
7TUH
 
 | | Crystal structure of anti-tapasin PaSta2-Fab | | Descriptor: | PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUD
 
 | | Crystal structure of HLA-B*44:05 (T73C) with 6mer EEFGRC and dipeptide GL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, EEFGRC peptide, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
6PUJ
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-3`OH-Propyl-5-OP-U | | Descriptor: | 6-[(3-hydroxypropyl)amino]-5-[(E)-(2-oxopropylidene)amino]pyrimidine-2,4(1H,3H)-dione, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
7TUG
 
 | | Crystal structure of Tapasin in complex with PaSta2-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7QHL
 
 | | Crystal structure of Cyclin-dependent kinase 2/cyclin A in complex with 3,5,7-Substituted pyrazolo[4,3-d]pyrimidine inhibitor 24 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-amino-1-ethyl)thio-3-cyclobutyl-7-[4-(pyrazol-1-yl)benzyl]amino-1(2)H-pyrazolo[4,3-d]pyrimidine, Cyclin-A2, ... | | Authors: | Djukic, S, Skerlova, J, Rezacova, P. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3,5,7-Substituted Pyrazolo[4,3- d ]Pyrimidine Inhibitors of Cyclin-Dependent Kinases and Cyclin K Degraders.
J.Med.Chem., 65, 2022
|
|
6GXE
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor JS14 | | Descriptor: | 3-[2-[2-[2-[2-(aminomethyloxy)ethoxy]ethoxy]ethoxy]ethoxy]-~{N}-[4-[4-[(4-sulfamoylphenyl)carbamoylamino]phenoxy]butyl]propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inhibitor-Polymer Conjugates as a Versatile Tool for Detection and Visualization of Cancer-Associated Carbonic Anhydrase Isoforms
Acs Omega, 2019
|
|
7TUE
 
 | | Crystal structure of Tapasin in complex with HLA-B*44:05 (T73C) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B alpha chain, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUF
 
 | | Crystal structure of Tapasin in complex with PaSta1-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta1 Fab heavy chain, PaSta1 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUC
 
 | | Crystal structure of HLA-B*44:05 (T73C) with 9mer EEFGRAFSF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
