5Y5W
 
 | |
8SRI
 
 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, closed state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8AKL
 
 | | Acyl-enzyme complex of meropenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2S,3R,4R)-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl-3-methyl-2-[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8ALZ
 
 | | Cryo-EM structure of ASCC3 in complex with ASC1 | | Descriptor: | Activating signal cointegrator 1, Activating signal cointegrator 1 complex subunit 3, ZINC ION | | Authors: | Jia, J, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of ASCC3 in complex with ASC1
Nat Commun, 2023
|
|
8SRH
 
 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRG
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, Adenosine monophosphate, and Ribose-5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
6P48
 
 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6VGU
 
 | |
8SRF
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, ADP-ribose, Adenosine monophosphate, and Ribose-5-phosphate, closed state | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRD
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
6UUF
 
 | | Crystal structure of a Nudix Hydrolase from M. Smegmatis, RenU | | Descriptor: | Nudix Hydrolase, RenU | | Authors: | Wright, K.M, Yoder, J, Shoemaker, S, Hernandez, A, Iheanacho, A, Marques, I, Amzel, M.L, Gabelli, S.B. | | Deposit date: | 2019-10-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of RenU
To Be Published
|
|
7X5L
 
 | | Tir-dsDNA complex, the initial binding state | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*AP*TP*TP*AP*A)-3'), Flax rust resistance protein | | Authors: | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | Deposit date: | 2022-03-04 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
3JZG
 
 | | Structure of EED in complex with H3K27me3 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6PI5
 
 | | The evolving story of AtzT, a periplasmic binding protein | | Descriptor: | Atrazine periplasmic binding protein, DIMETHYL SULFOXIDE, GUANINE | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | Deposit date: | 2019-06-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8SNB
 
 | |
8AAC
 
 | |
7QB1
 
 | | PPARg in complex with inhibitor | | Descriptor: | 3-[(3,4-dichlorophenyl)methyl]-4-oxidanylidene-6-phenoxy-phthalazine-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Petersen, J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery by Virtual Screening of an Inhibitor of CDK5-Mediated PPAR gamma Phosphorylation.
Acs Med.Chem.Lett., 13, 2022
|
|
3ONJ
 
 | |
2MZJ
 
 | |
4L8B
 
 | | Crystal structure of the H2Db in complex with the NP-N5H peptide | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, H-2 class I histocompatibility antigen, ... | | Authors: | Rossjohn, J, Gras, S. | | Deposit date: | 2013-06-16 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acute emergence and reversion of influenza A virus quasispecies within CD8(+) T cell antigenic peptides.
Nat Commun, 4, 2013
|
|
7X5M
 
 | | Tir-dsDNA complex, the initial binding state | | Descriptor: | 2',3'- cyclic AMP, DNA (5'-D(P*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*AP*TP*TP*TP*A)-3'), ... | | Authors: | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
8ADA
 
 | | Crystal structure of ClpC2 N-terminal domain | | Descriptor: | Uncharacterized protein Rv2667 | | Authors: | Taylor, G, Cui, H.J, Leodolter, J, Giese, C, Weber-Ban, E. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | ClpC2 protects mycobacteria against a natural antibiotic targeting ClpC1-dependent protein degradation.
Commun Biol, 6, 2023
|
|
8SOY
 
 | |
8AE7
 
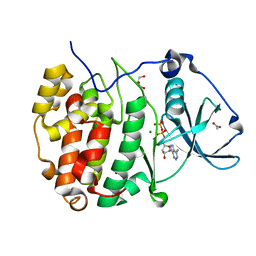 | | The strucuture of Compound 15 bound to CK2alpha | | Descriptor: | 2-[5,6-bis(bromanyl)-1H-indazol-3-yl]ethanenitrile, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Atkinson, E, Frances, N, Iegre, J, Venkitaraman, A, Hyvonen, M, Spring, D. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
2N1R
 
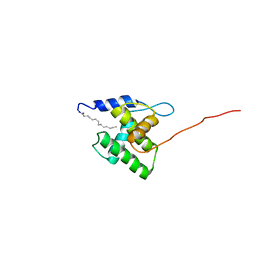 | | NMR Structure of the Myristylated Feline Immunodeficiency Virus Matrix Protein | | Descriptor: | Matrix protein p15 | | Authors: | Brown, L.A, Cox, C, Button, R.J, Baptiste, J, Bahlow, K, Spurrier, V, Luttge, B.G, Kuo, L, Freed, E.O, Summers, M.F, Kyser, J, Summers, H.R. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the myristylated feline immunodeficiency virus matrix protein.
Viruses, 7, 2015
|
|
