4GMP
 
 | |
7JP3
 
 | | Des-B29,B30-insulin | | Descriptor: | CHLORIDE ION, Insulin B chain,Insulin A chain, PHENOL, ... | | Authors: | Yoder, J, Weiss, M.A, DiMarchi, R, Zaykov, A. | | Deposit date: | 2020-08-07 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Des-B29,B30-insulin
To Be Published
|
|
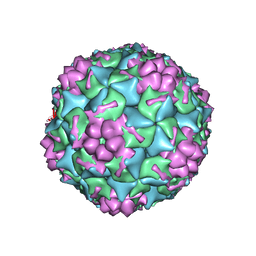
6MBA
 
 | | Crystal Structure of Human Nav1.4 CTerminal Domain in Complex with apo Calmodulin | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONATE ION, ... | | Authors: | Yoder, J, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Ca2+-dependent regulation of sodium channels NaV1.4 and NaV1.5 is controlled by the post-IQ motif.
Nat Commun, 10, 2019
|
|
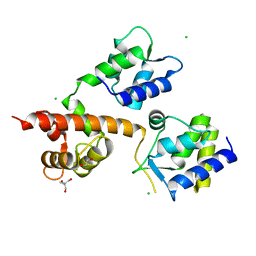
4GB3
 
 | | Human coxsackievirus B3 strain RD coat protein | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, coat protein 1, ... | | Authors: | Yoder, J.D, Hafenstein, S. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface.
J.Virol., 86, 2012
|
|
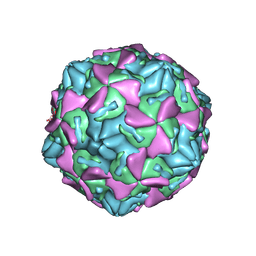
2B4I
 
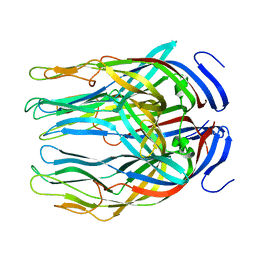 | |
2B4H
 
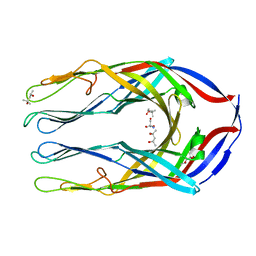 | |
6MC9
 
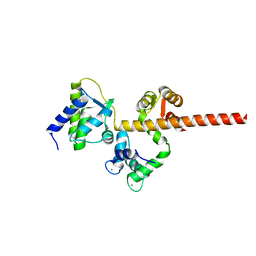 | |
3J24
 
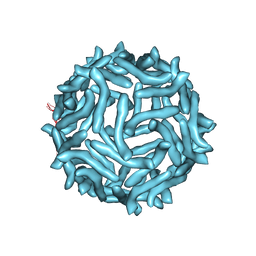 | | CryoEM reconstruction of complement decay-accelerating factor | | Descriptor: | Complement decay-accelerating factor | | Authors: | Yoder, J.D, Hafenstein, S.H. | | Deposit date: | 2012-08-17 | | Release date: | 2012-09-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface.
J.Virol., 86, 2012
|
|
5JFT
 
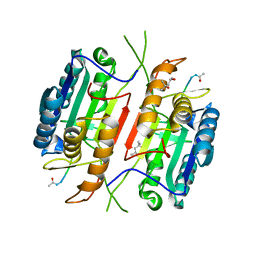 | | Zebra Fish Caspase-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACE-ASP-GLU-VAL-ASK, ... | | Authors: | Tucker, M.B, MacKenzie, S.H, Maciag, J.J, Dirscherl, H, Swartz, P.D, Yoder, J.A, Hamilton, P.T, Clark, A.C. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Phage display and structural studies reveal plasticity in substrate specificity of caspase-3a from zebrafish.
Protein Sci., 25, 2016
|
|
1G55
 
 | | Structure of human DNMT2, an enigmatic DNA methyltransferase homologue | | Descriptor: | BETA-MERCAPTOETHANOL, DNA CYTOSINE METHYLTRANSFERASE DNMT2, GLYCEROL, ... | | Authors: | Dong, A, Yoder, J.A, Zhang, X, Zhou, L, Bestor, T.H, Cheng, X. | | Deposit date: | 2000-10-30 | | Release date: | 2001-01-17 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA.
Nucleic Acids Res., 29, 2001
|
|
6UUF
 
 | | Crystal structure of a Nudix Hydrolase from M. Smegmatis, RenU | | Descriptor: | Nudix Hydrolase, RenU | | Authors: | Wright, K.M, Yoder, J, Shoemaker, S, Hernandez, A, Iheanacho, A, Marques, I, Amzel, M.L, Gabelli, S.B. | | Deposit date: | 2019-10-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of RenU
To Be Published
|
|
6VEA
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Glycine | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCINE, Glutamate receptor 3.2, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6VE8
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
