4TQN
 
 | |
8C0B
 
 | | CryoEM structure of Aspergillus nidulans UTP-glucose-1-phosphate uridylyltransferase | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Han, X, D Angelo, C, Otamendi, A, Cifuente, J.O, de Astigarraga, E, Ochoa-Lizarralde, B, Grininger, M, Routier, F.H, Guerin, M.E, Fuehring, J, Etxebeste, O, Connell, S.R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | CryoEM analysis of the essential native UDP-glucose pyrophosphorylase from Aspergillus nidulans reveals key conformations for activity regulation and function.
Mbio, 14, 2023
|
|
6PWW
 
 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
8CII
 
 | | Delta-RBD complex with BA.2-07 fab, SARS1-34 fab and C1 nanobody | | Descriptor: | BA.2-07 fab Heavy Chain, BA.2-07 fab Light Chain, C1 nanobody, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Potent cross-reactive mAbs from BA.4/5 breakthrough infection
To Be Published
|
|
7UGB
 
 | |
8BYH
 
 | |
4J32
 
 | | Structure of the effector - immunity system Tae4 / Tai4 from Salmonella typhimurium | | Descriptor: | 2-ETHOXYETHANOL, CITRATE ANION, Putative cytoplasmic protein, ... | | Authors: | Benz, J, Reinstein, J, Meinhart, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tae4/Tai4 from Salmonella typhimurium.
Plos One, 8, 2013
|
|
5WWJ
 
 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1).
To Be Published
|
|
2FQN
 
 | |
4J0M
 
 | | Crystal structure of BRL1 (LRR) in complex with brassinolide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, ... | | Authors: | Chai, J, She, J, Han, Z, Zhou, B. | | Deposit date: | 2013-01-31 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for differential recognition of brassinolide by its receptors
Protein Cell, 4, 2013
|
|
8BZV
 
 | |
4EL1
 
 | | Crystal structure of oxidized hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
8C7Z
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2308 | | Descriptor: | 1,2-ETHANEDIOL, 9-piperazin-1-yl-4-(3,4,5-trimethoxyphenyl)-5,6-dihydro-[1]benzoxepino[5,4-c]pyridine, AMMONIUM ION, ... | | Authors: | Cros, J, Williams, E.P, Sweeney, M.N, Smil, D, Gonzalez-Alvarez, H, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery of Conformationally Constrained ALK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
6Q1Q
 
 | |
6Q2F
 
 | | Structure of Rhamnosidase from Novosphingobium sp. PP1Y | | Descriptor: | Glycoside hydrolase family protein, SODIUM ION | | Authors: | Terry, B, Ha, J, Izzo, V, Sazinsky, M.H. | | Deposit date: | 2019-08-07 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.20000076 Å) | | Cite: | The crystal structure and insight into the substrate specificity of the alpha-L rhamnosidase RHA-P from Novosphingobium sp. PP1Y.
Arch.Biochem.Biophys., 679, 2019
|
|
8C7W
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2304 | | Descriptor: | 1,2-ETHANEDIOL, 6-methyl-9-piperazin-1-yl-4-(3,4,5-trimethoxyphenyl)-5,7-dihydropyrido[4,3-d][2]benzazepine, Activin receptor type I, ... | | Authors: | Cros, J, Williams, E.P, Sweeney, M.N, Smil, D, Gonzalez-Alvarez, H, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Conformationally Constrained ALK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
7JW6
 
 | | Crystal structure of Drosophila Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2O1Z
 
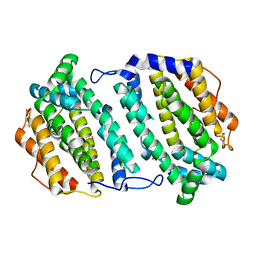 | | Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155) | | Descriptor: | FE (III) ION, Ribonucleotide Reductase Subunit R2, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Tempel, W, Qiu, W, Lew, J, Wernimont, A.K, Lin, Y.H, Hassanali, A, Melone, M, Zhao, Y, Nordlund, P, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155)
To be Published
|
|
8BYK
 
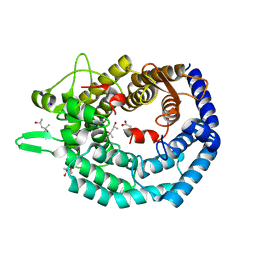 | | The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Knospe, C.V, Kamel, M, Spitz, O, Hoeppner, A, Galle, S, Reiners, J, Kedrov, A, Smits, S.H, Schmitt, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases.
Front Microbiol, 13, 2022
|
|
1A94
 
 | | STRUCTURAL BASIS FOR SPECIFICITY OF RETROVIRAL PROTEASES | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | Authors: | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 1998-04-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
2FSP
 
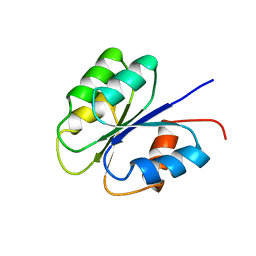 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
6PSR
 
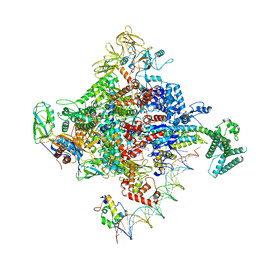 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
5UY8
 
 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | 5-[(5S)-5-ethyl-5-methyl-6-oxo-1,4,5,6-tetrahydropyridin-3-yl]-N-(6-fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)thiophene-2-sulfonamide, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, ... | | Authors: | Wang, J, Wang, Y, Fales, K.R, Atwell, S, Clawson, D. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
8CKS
 
 | | Crystal structure of Human Serum Albumin in complex with FESAN | | Descriptor: | 3,3'-commo-bis(1,2-dicarba-3-ferra-closo-dodecaborane), DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Dolot, R.M, Kaniowski, D, Ebenryter-Olbinska, K, Szczupak, P, Suwara, J, Nawrot, B.C. | | Deposit date: | 2023-02-16 | | Release date: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Serum Albumin in complex with FESAN
To Be Published
|
|
5UZ0
 
 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, MAGNESIUM ION, ... | | Authors: | Atwell, S, Wang, Y, Fales, K.R, Clawson, D, Wang, J. | | Deposit date: | 2017-02-24 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
