1EFG
 
 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR G COMPLEXED WITH GDP, AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1994-10-17 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
5ZB4
 
 | | Crystal structure of thymidylate kinase in complex with ADP and TMP from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chaudhary, S.K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into product release dynamics through structural analyses of thymidylate kinase.
Int. J. Biol. Macromol., 123, 2018
|
|
8F1I
 
 | | SigN RNA polymerase early-melted intermediate bound to mismatch fragment dhsU36mm1 (-12T) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1J
 
 | | SigN RNA polymerase early-melted intermediate bound to mismatch DNA fragment dhsU36mm2 (-12A) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5KLP
 
 | | Crystal structure of HopZ1a in complex with IP6 | | Descriptor: | CITRIC ACID, INOSITOL HEXAKISPHOSPHATE, Orf34 | | Authors: | Zhang, Z.-M, Song, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-08-10 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8F1K
 
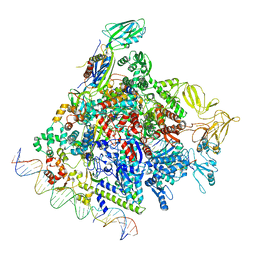 | | SigN RNA polymerase early-melted intermediate bound to full duplex DNA fragment dhsU36 (-12T) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WRI
 
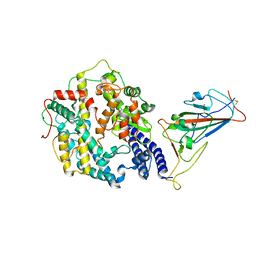 | | Cryo-EM structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-08 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
5KOR
 
 | | Arabidopsis thaliana fucosyltransferase 1 (FUT1) in complex with GDP and a xylo-oligossacharide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rocha, J, de Sanctis, D, Breton, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation.
Plant Cell, 28, 2016
|
|
5KPQ
 
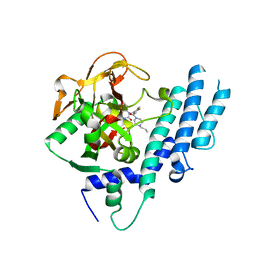 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-propyl-piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5KQ4
 
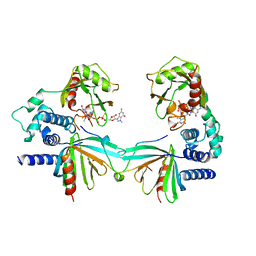 | | Crystal structure of S. pombe Dcp1/Dcp2 in complex with H. sapiens PNRC2 and synthetic cap analog | | Descriptor: | Proline-rich nuclear receptor coactivator 2, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] [[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-oxidanyl-phosphoryl] hydrogen phosphate, mRNA decapping complex subunit 2, ... | | Authors: | Mugridge, J.S, Ziemniak, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of mRNA-cap recognition by Dcp1-Dcp2.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5KQM
 
 | | Co-crystal structure of LMW-PTP in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Wang, J, Zhang, Z.-Y, Yu, Z.-H. | | Deposit date: | 2016-07-06 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Inhibition of low molecular weight protein tyrosine phosphatase by an induced-fit mechanism.
J.Med.Chem., 2016
|
|
7WSK
 
 | | Crystal structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with civet ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Huang, B, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
6ELL
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
5KRF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Dynamic WAY derivative, 1a | | Descriptor: | 4-[1-methyl-7-(trifluoromethyl)indazol-3-yl]benzene-1,3-diol, Estrogen receptor, KHKILHRLLQDSSS Peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5KVA
 
 | | Crystal Structure of sorghum caffeoyl-CoA O-methyltransferase (CCoAOMT) | | Descriptor: | CALCIUM ION, S-ADENOSYLMETHIONINE, caffeoyl-CoA O-methyltransferase | | Authors: | Walker, A.M, Sattler, S.A, Regner, M, Jones, J.P, Ralph, J, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | The Structure and Catalytic Mechanism of Sorghum bicolor Caffeoyl-CoA O-Methyltransferase.
Plant Physiol., 172, 2016
|
|
5KVR
 
 | | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073 | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073
To Be Published
|
|
8EQ5
 
 | | Crystal structure of the N-terminal kinase domain of RSK2 in complex with SPRED2 (131-160) | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, Ribosomal protein S6 kinase alpha-3, Sprouty-related, ... | | Authors: | Bonsor, D.A, Lopez, J, McCormick, F, Simanshu, D.K. | | Deposit date: | 2022-10-07 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The ribosomal S6 kinase 2 (RSK2)-SPRED2 complex regulates the phosphorylation of RSK substrates and MAPK signaling.
J.Biol.Chem., 299, 2023
|
|
5KVB
 
 | |
3SWV
 
 | | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A | | Descriptor: | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-14 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BrefeldinA-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A
To be Published
|
|
3LNX
 
 | | Second PDZ domain from human PTP1E | | Descriptor: | IODIDE ION, THIOCYANATE ION, Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Zhang, J, Chang, A, Ke, H, Phillips Jr, G.N, Lee, A.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-02-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E.
Biochemistry, 49, 2010
|
|
5KW2
 
 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
8WKP
 
 | |
8VLX
 
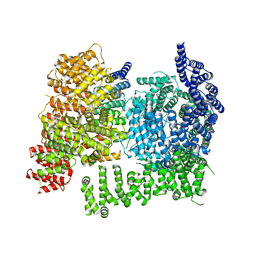 | | HTT in complex with HAP40 and a small molecule. | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin, N-[(1S,2R)-2-benzylcyclopentyl]-N'-{1-[(1S)-1-(pyridin-4-yl)ethyl]piperidin-4-yl}urea | | Authors: | Poweleit, N, Boudet, J, Doherty, E. | | Deposit date: | 2024-01-12 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Discovery of a Small Molecule Ligand to the Huntingtin/HAP40 complex
To Be Published
|
|
8W7N
 
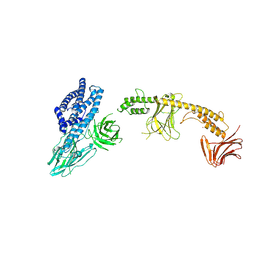 | | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis | | Descriptor: | Pesticidal crystal protein Cry1Aa, UNKNOWN ATOM OR ION | | Authors: | Tanaka, J, Abe, S, Hayakawa, T, Kojima, M, Yamashita, K, Hirata, K, Ueno, T. | | Deposit date: | 2023-08-31 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 685, 2023
|
|
5YVT
 
 | |
