6FGS
 
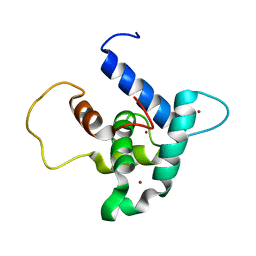 | | Solution structure of p300Taz2-p73TA1 | | Descriptor: | Histone acetyltransferase p300,Tumor protein p73, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
7WQH
 
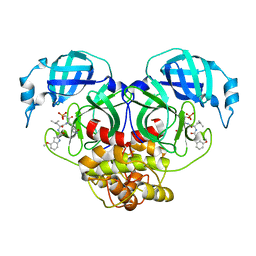 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7DYS
 
 | |
7WQJ
 
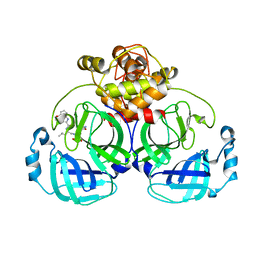 | | Crystal structure of MERS main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
6EZX
 
 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-17-oxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-[1-(iminomethyl)cyclopropyl]-5-oxidanylidene-17-oxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide, Cathepsin L1 | | Authors: | Banner, D.W, Benz, J, Kuglstatter, A. | | Deposit date: | 2017-11-16 | | Release date: | 2018-04-11 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
7WQL
 
 | | Bovin Beta-lactoglobulin binding with zinc ions | | Descriptor: | Beta-lactoglobulin, ZINC ION | | Authors: | Li, T, Ma, J, Zang, J, Zhao, G, Zhang, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Zinc binding strength of proteins dominants zinc uptake in Caco-2 cells.
Rsc Adv, 12, 2022
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
4MTJ
 
 | | Structure of the b12-independent glycerol dehydratase with 1,2-propanediol bound | | Descriptor: | B12-independent glycerol dehydratase, S-1,2-PROPANEDIOL | | Authors: | LaMattina, J, Wright, A.V, Demick, J, Soucaille, P, Lanzilotta, W.N. | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | When Computational Chemistry and Modern Software Get It Right; New Insight Into the Mechanism of a Glycyl Radical Enzyme
To be Published
|
|
3DFR
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI AND LACTOBACILLUS CASEI DIHYDROFOLATE REDUCTASE REFINED AT 1.7 ANGSTROMS RESOLUTION. I. GENERAL FEATURES AND BINDING OF METHOTREXATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Filman, D.J, Matthews, D.A, Bolin, J.T, Kraut, J. | | Deposit date: | 1982-06-25 | | Release date: | 1982-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate.
J.Biol.Chem., 257, 1982
|
|
6F81
 
 | | AKR1B1 at 0.75 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
7EJH
 
 | | Crystal structure of KRED mutant-F147L/L153Q/Y190P/L199A/M205F/M206F and 2-hydroxyisoindoline-1,3-dione complex | | Descriptor: | 2-oxidanylisoindole-1,3-dione, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72883928 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7EJJ
 
 | | Crystal structure of KRED F147L/L153Q/Y190P variant and methyl methacrylate complex | | Descriptor: | 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.80000663 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7EJI
 
 | | Crystal structure of KRED F147L/L153Q/Y190P/L199A/M205F/M206F variant and methyl methacrylate complex | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxidanylisoindole-1,3-dione, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.560016 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
6ETD
 
 | |
4L43
 
 | | Crystal structures of human p70S6K1-T389A (form I) | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
4KVH
 
 | | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747 | | Descriptor: | BROMIDE ION, CACODYLATE ION, FORMIC ACID, ... | | Authors: | Chang, C, Holowicki, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747
To be Published
|
|
2WGH
 
 | | Human Ribonucleotide reductase R1 subunit (RRM1) in complex with dATP and Mg. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Welin, R.M, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh-Nielsen, T, Kotzsch, A, Kotenyova, T, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wisniewska, M, Nordlund, P. | | Deposit date: | 2009-04-19 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Allosteric Regulation of Human Ribonucleotide Reductase by Nucleotide-Induced Oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6EX6
 
 | | The GH127, Beta-arabinofuranosidase, BT3674 | | Descriptor: | Six-hairpin glycosidase, ZINC ION | | Authors: | Munoz-Munoz, J, Gilbert, H.J. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The cellular location of endo-acting galactanases confers keystone or recipient status to arabinogalactan degrading organisms of the human gut microbiota
To Be Published
|
|
7W0V
 
 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4LAN
 
 | | Crystal structure of Cordyceps militaris IDCase H195A mutant | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
6F3N
 
 | |
2WCY
 
 | | NMR solution structure of factor I-like modules of complement C7. | | Descriptor: | COMPLEMENT COMPONENT C7 | | Authors: | Phelan, M.M, Thai, C.T, Soares, D.C, Ogata, R.T, Barlow, P.N, Bramham, J. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Factor I-Like Modules from Complement C7 Reveals a Pair of Follistatin Domains in Compact Pseudosymmetric Arrangement.
J.Biol.Chem., 284, 2009
|
|
4L3L
 
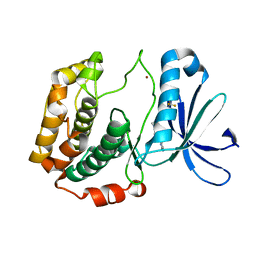 | | Crystal structures of human p70S6K1 kinase domain (Zinc anomalous) | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein, ZINC ION | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
2VCI
 
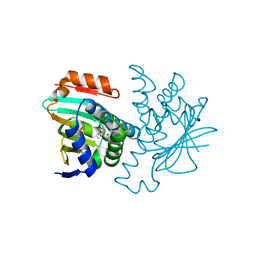 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-[2,4-DIHYDROXY-5-(1-METHYLETHYL)PHENYL]-N-ETHYL-4-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]ISOXAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
2VCJ
 
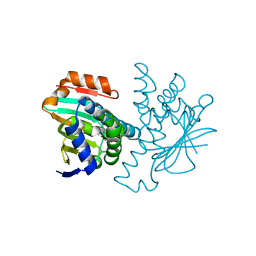 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-(5-chloro-2,4-dihydroxyphenyl)-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]isoxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
