6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6NE9
 
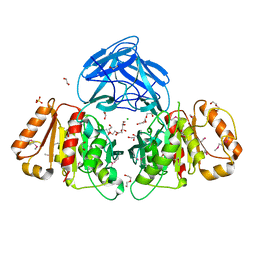 | | Bacteroides intestinalis acetyl xylan esterase (BACINT_01039) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Pereira, G.V, Cann, I. | | Deposit date: | 2018-12-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Degradation of complex arabinoxylans by human colonic Bacteroidetes
Nat Commun, 2021
|
|
1T32
 
 | | A Dual Inhibitor of the Leukocyte Proteases Cathepsin G and Chymase with Therapeutic Efficacy in Animals Models of Inflammation | | Descriptor: | 2-[3-({METHYL[1-(2-NAPHTHOYL)PIPERIDIN-4-YL]AMINO}CARBONYL)-2-NAPHTHYL]-1-(1-NAPHTHYL)-2-OXOETHYLPHOSPHONIC ACID, Cathepsin G, SULFATE ION | | Authors: | de Garavilla, L, Greco, M.N, Giardino, E.C, Wells, G.I, Haertlein, B.J, Kauffman, J.A, Corcoran, T.W, Derian, C.K, Eckardt, A.J, Abraham, W.M, Sukumar, N, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E, Andrade-Gordon, P, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2004-04-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel, potent dual inhibitor of the leukocyte proteases cathepsin G and chymase: molecular mechanisms and anti-inflammatory activity in vivo.
J.Biol.Chem., 280, 2005
|
|
2PFF
 
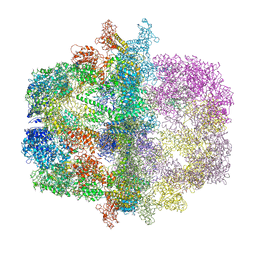 | | Structural Insights of Yeast Fatty Acid Synthase | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, Tail protein | | Authors: | Xiong, Y, Lomakin, I.B, Steitz, T.A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Insights of Yeast Fatty Acid Synthase
Cell(Cambridge,Mass.), 129, 2007
|
|
1SW8
 
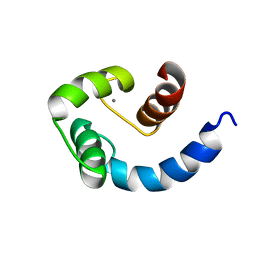 | | Solution structure of the N-terminal domain of Human N60D calmodulin refined with paramagnetism based strategy | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Bertini, I, Del Bianco, C, Gelis, I, Katsaros, N, Luchinat, C, Parigi, G, Peana, M, Provenzani, A, Zoroddu, M.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally exploring the conformational space sampled by domain reorientation in calmodulin
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3I3Z
 
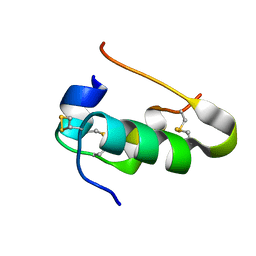 | | Human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Timofeev, V.I, Bezuglov, V.V, Miroshnikov, K.A, Cuprov-Netochin, R.N, Samigina, V.R, Kuranova, I.P. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray investigation of gene-engineered human insulin crystallized from a solution containing polysialic acid.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5ZAB
 
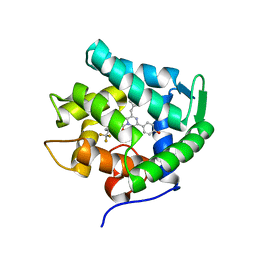 | | Crystal structure of cf3-aequorin | | Descriptor: | (2S)-8-benzyl-2-hydroperoxy-6-(4-hydroxyphenyl)-2-{[4-(trifluoromethyl)phenyl]methyl}imidazo[1,2-a]pyrazin-3(2H)-one, Aequorin-2 | | Authors: | Inouye, S, Tomabechi, Y, Sekine, S.I, Shirouzu, M, Hosoya, T. | | Deposit date: | 2018-02-07 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Slow luminescence kinetics of semi-synthetic aequorin: expression, purification and structure determination of cf3-aequorin.
J. Biochem., 164, 2018
|
|
1PXO
 
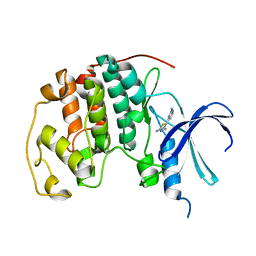 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR [4-(2-Amino-4-methyl-thiazol-5-yl)-pyrimidin-2-yl]-(3-nitro-phenyl)-amine | | Descriptor: | Cell division protein kinase 2, [4-(2-AMINO-4-METHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YL]-(3-NITRO-PHENYL)-AMINE | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
5EDF
 
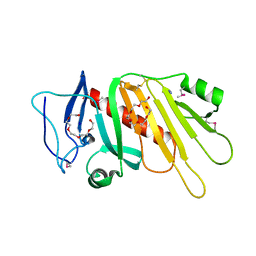 | | Crystal structure of the selenomethionine-substituted iron-regulated protein FrpD from Neisseria meningitidis | | Descriptor: | AZIDE ION, FrpC operon protein, HEXAETHYLENE GLYCOL, ... | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
7MD7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with triphenylphosphonium analog of chloramphenicol CAM-C4-TPP and protein Y (YfiA) at 2.80A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Pavlova, J.A, Lukianov, D.A, Tereshchenkov, A.G, Makarov, G.I, Khairullina, Z.Z, Tashlitsky, V.N, Paleskava, A, Konevega, A.L, Bogdanov, A.A, Osterman, I.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2021-04-03 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Triphenylphosphonium Analog of Chloramphenicol upon the Bacterial Ribosome.
Antibiotics, 10, 2021
|
|
1PXM
 
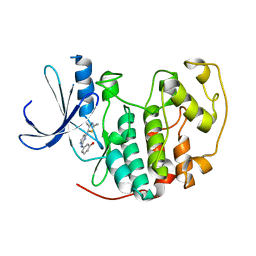 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 3-[4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-ylamino]-phenol | | Descriptor: | 3-[4-(2,4-DIMETHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-PHENOL, Cell division protein kinase 2 | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1Q0K
 
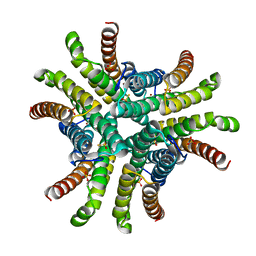 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the thiosulfate-reduced state | | Descriptor: | NICKEL (II) ION, SULFATE ION, Superoxide dismutase [Ni], ... | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5VFF
 
 | |
2JSW
 
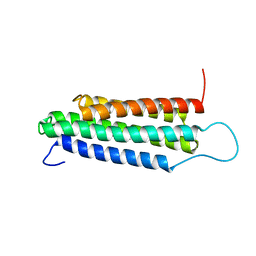 | | Solution Structure of the R13 Domain of Talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Critchley, D.R.C, Barsukov, I.L. | | Deposit date: | 2007-07-17 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the C-terminal actin-binding domain of talin.
Embo J., 27, 2007
|
|
2FX7
 
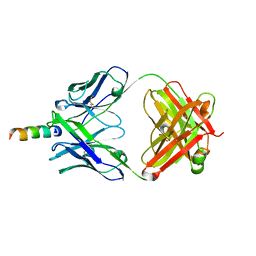 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with a 16-residue peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein (GP41), GLYCEROL | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
2PZB
 
 | | NAD+ Synthetase from Bacillus anthracis | | Descriptor: | NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | McDonald, H.M, Pruett, P.S, Deivanayagam, C, Protasevich, I.I, Carson, W.M, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural adaptation of an interacting non-native C-terminal helical extension revealed in the crystal structure of NAD(+) synthetase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3P4U
 
 | | Crystal structure of active caspase-6 in complex with Ac-VEID-CHO inhibitor | | Descriptor: | Ac-VEID-CHO inhibitor, Caspase-6 | | Authors: | Mueller, I, Lamers, M.B.A.C, Ritchie, A.J, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2010-10-07 | | Release date: | 2011-09-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of active and inhibitor-bound human Casp6
To be Published
|
|
1N0T
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
3P68
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | GOLD 3+ ION, Lysozyme C | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
2Y69
 
 | | Bovine heart cytochrome c oxidase re-refined with molecular oxygen | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, CHOLIC ACID, ... | | Authors: | Kaila, V.R.I, Oksanen, E, Goldman, A, Verkhovsky, M.I, Sundholm, D, Wikstrom, M. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Combined Quantum Chemical and Crystallographic Study on the Oxidized Binuclear Center of Cytochrome C Oxidase.
Biochim.Biophys.Acta, 1807, 2011
|
|
2ARJ
 
 | | CD8alpha-alpha in complex with YTS 105.18 Fab | | Descriptor: | T-cell surface glycoprotein CD8 alpha chain, YTS 105.18 antigen binding region Heavy chain, YTS 105.18 antigen binding region Light chain | | Authors: | Shore, D.A, Teyton, L, Dwek, R.A, Rudd, P.M, Wilson, I.A. | | Deposit date: | 2005-08-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the TCR co-receptor CD8alphaalpha in complex with monoclonal antibody YTS 105.18 Fab fragment at 2.88 A resolution.
J.Mol.Biol., 358, 2006
|
|
7K76
 
 | |
6EC0
 
 | | Crystal structure of the wild-type heterocomplex between coil 1B domains of human intermediate filament proteins keratin 1 (KRT1) and keratin 10 (KRT10) | | Descriptor: | CADMIUM ION, COBALT (II) ION, Keratin 1, ... | | Authors: | Eldirany, S.A, Lomakin, I.B, Bunick, C.G. | | Deposit date: | 2018-08-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly.
Embo J., 38, 2019
|
|
1Q0D
 
 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the oxidized state | | Descriptor: | NICKEL (III) ION, SULFATE ION, Superoxide dismutase [Ni] | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2CDF
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5E) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
