8WKX
 
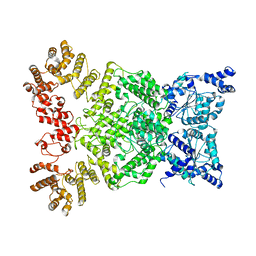 | | Cryo-EM structure of DSR2 | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
8WKT
 
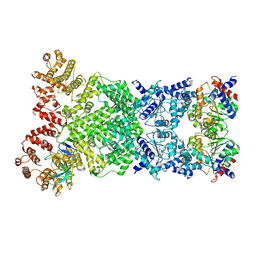 | | Cryo-EM structure of DSR2-DSAD1 complex | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
5HC4
 
 | | Structure of esterase Est22 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5DWD
 
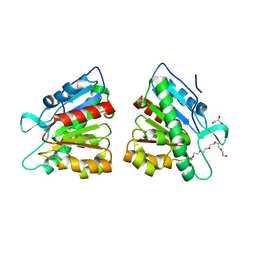 | | Crystal structure of esterase PE8 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Esterase, GLYCEROL | | Authors: | Li, J, Huang, J. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of esterase PE8
To Be Published
|
|
8FHD
 
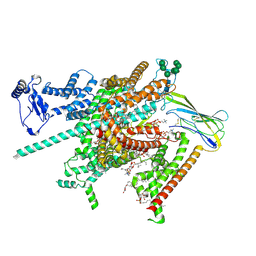 | | Cryo-EM structure of human voltage-gated sodium channel Nav1.6 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (5E,17R,20S)-23-amino-20-hydroxy-14,20-dioxo-15,19,21-trioxa-20lambda~5~-phosphatricos-5-en-17-yl hexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of human voltage-gated sodium channel Na v 1.6.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5HC5
 
 | |
5HC0
 
 | | Structure of esterase Est22 with p-nitrophenol | | Descriptor: | ACETIC ACID, GLYCEROL, Lipolytic enzyme, ... | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5C13
 
 | |
5HC3
 
 | | The structure of esterase Est22 | | Descriptor: | GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC2
 
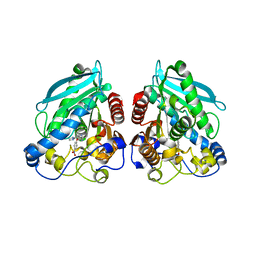 | |
4E9A
 
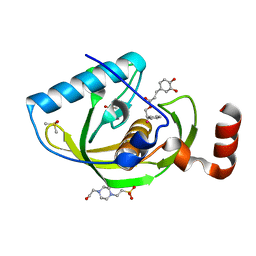 | | Structure of Peptide Deformylase form Helicobacter Pylori in complex with inhibitor | | Descriptor: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COBALT (II) ION, ... | | Authors: | Cui, K, Zhu, L, Lu, W, Huang, J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Identification of Novel Peptide Deformylase Inhibitors from Natural Products
To be Published
|
|
4CC5
 
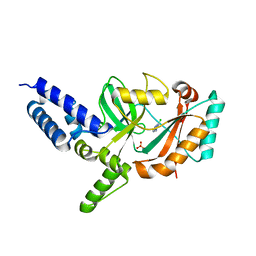 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
4CC6
 
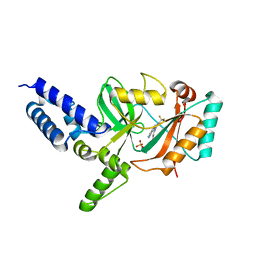 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-{[2-(1H-pyrazolo[3,4-c]pyridin-3-yl)-6-(trifluoromethyl)pyridin-4-yl]amino}ethanol, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
4E9B
 
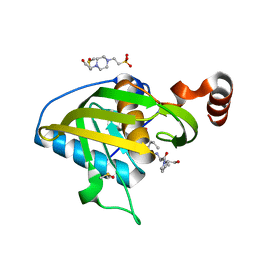 | | Structure of Peptide Deformylase form Helicobacter Pylori in complex with actinonin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACTINONIN, COBALT (II) ION, ... | | Authors: | Cui, K, Zhu, L, Lu, W, Huang, J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-04-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Novel Peptide Deformylase Inhibitors from Natural Products
To be Published
|
|
4IG4
 
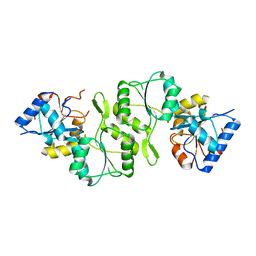 | | Crystal structure of single mutant thermostable NPPase (N86S) from Geobacillus stearothermophilus | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Wang, F, Huang, J, Qiu, R, Yang, Z, Wang, Y, Gong, W, Ji, C. | | Deposit date: | 2012-12-16 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of thermostable NPPase from Geobacillus stearothermophilus
To be Published
|
|
4KN8
 
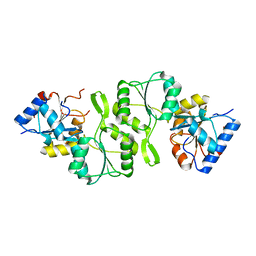 | | Crystal structure of Bs-TpNPPase | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Wang, F, Huang, J, Gong, W, Ji, C. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal Structure of Thermostable p-nitrophenylphosphatase from Bacillus Stearothermophilus (Bs-TpNPPase)
PROTEIN PEPT.LETT., 21, 2014
|
|
4IFT
 
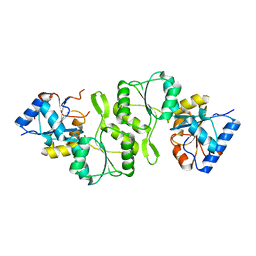 | | Crystal structure of double mutant thermostable NPPase from Geobacillus stearothermophilus | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Huang, J, Wang, F, Qiu, R, Wang, Y, Ji, C. | | Deposit date: | 2012-12-15 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of thermostable NPPase from Geobacillus stearothermophilus
To be Published
|
|
5GP4
 
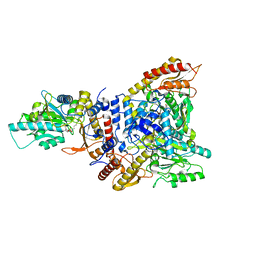 | | Lactobacillus brevis CGMCC 1306 Glutamate decarboxylase | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mei, L, Huang, J. | | Deposit date: | 2016-07-31 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Lactobacillus brevis CGMCC 1306 glutamate decarboxylase: Crystal structure and functional analysis.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
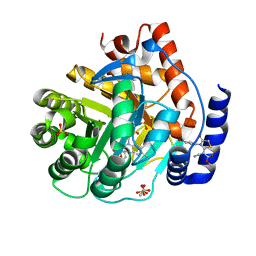
5H2Z
 
 | |
3TYH
 
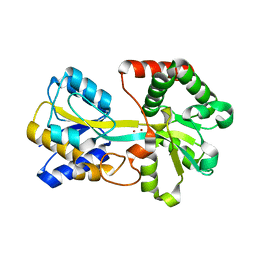 | | Crystal structure of oxo-cupper clusters binding to ferric binding protein from Neisseria gonorrhoeae | | Descriptor: | COPPER (II) ION, FbpA protein | | Authors: | Chen, W.J, Wang, H.F, Zhou, C.J, Ye, D.R, Huang, J, Tan, X.S, Zhong, W.Q. | | Deposit date: | 2011-09-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of oxo-cupper clusters binding to ferric binding protein from Neisseria gonorrhoeae
To be Published
|
|
3U4V
 
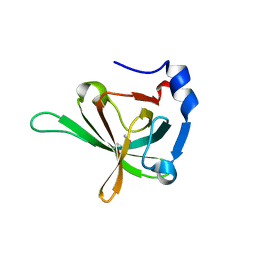 | |
3U4Z
 
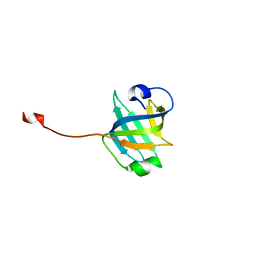 | |
3U50
 
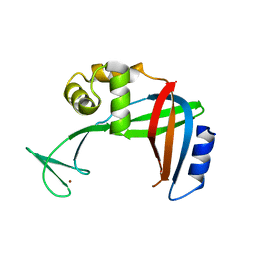 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 OB-C | | Descriptor: | Telomerase-associated protein 82, ZINC ION | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U58
 
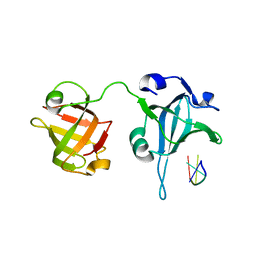 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 AB | | Descriptor: | DNA (5'-D(*GP*GP*GP*T)-3'), Tetrahymena Teb1 AB | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5ZG9
 
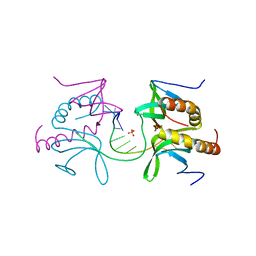 | | Crystal structure of MoSub1-ssDNA complex in phosphate buffer | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*G)-3'), MoSub1, PHOSPHATE ION | | Authors: | Zhao, Y, Huang, J, Liu, H, Yi, L, Wang, S, Zhang, X, Liu, J. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The effect of phosphate ion on the ssDNA binding mode of MoSub1, a Sub1/PC4 homolog from rice blast fungus.
Proteins, 87, 2019
|
|
