8XL6
 
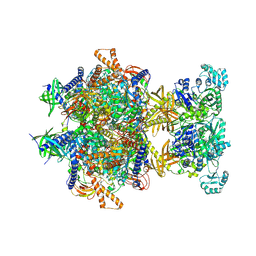 | | Structure of human 3-methylcrotonyl-CoA carboxylase at apo-state (MCC-Apo) | | 分子名称: | BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | 著者 | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | 登録日 | 2023-12-25 | | 公開日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.29 Å) | | 主引用文献 | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL7
 
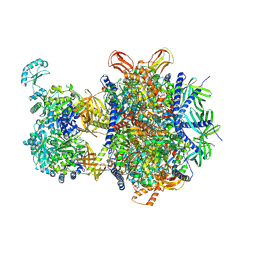 | | Structure of human 3-methylcrotonyl-CoA carboxylase in complex with acetyl-CoA (MCC-ACO) | | 分子名称: | ACETYL COENZYME *A, BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, ... | | 著者 | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | 登録日 | 2023-12-25 | | 公開日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL8
 
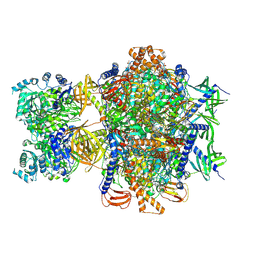 | | Structure of human 3-methylcrotonyl-CoA carboxylase in complex with propionyl-CoA (MCC-PCO) | | 分子名称: | BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | 著者 | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | 登録日 | 2023-12-25 | | 公開日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL4
 
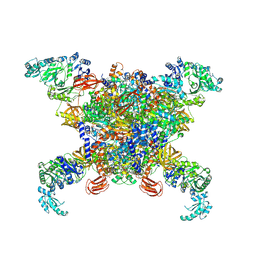 | | Structure of human propionyl-CoA carboxylase in complex with acetyl-CoA (PCC-ACO) | | 分子名称: | ACETYL COENZYME *A, BIOTIN, Propionyl-CoA carboxylase alpha chain, ... | | 著者 | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | 登録日 | 2023-12-25 | | 公開日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL5
 
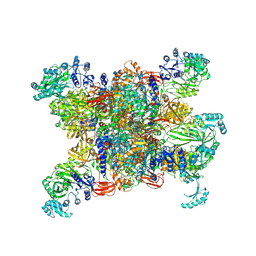 | | Structure of human propionyl-CoA carboxylase in complex with propionyl-CoA (PCC-PCO) | | 分子名称: | BIOTIN, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | 著者 | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | 登録日 | 2023-12-25 | | 公開日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
7QBZ
 
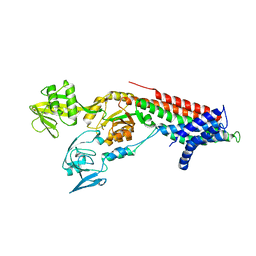 | | Crystal structure Cadmium translocating P-type ATPase | | 分子名称: | Cadmium translocating P-type ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | 著者 | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | 登録日 | 2021-11-21 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7QC0
 
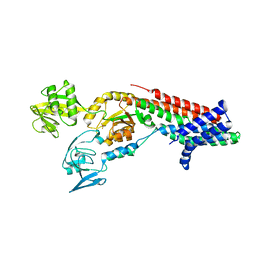 | | Crystal structure of Cadmium translocating P-type ATPase | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, Cadmium translocating P-type ATPase, MAGNESIUM ION | | 著者 | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | 登録日 | 2021-11-21 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
5KN7
 
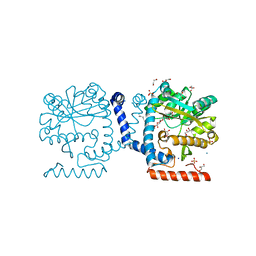 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | 著者 | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | 登録日 | 2016-06-27 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GPO
 
 | | The sensor domain structure of the zinc-responsive histidine kinase CzcS from Pseudomonas Aeruginosa | | 分子名称: | SULFATE ION, Sensor protein CzcS, ZINC ION | | 著者 | Wang, D, Chen, W.Z, Huang, S.Q, Liu, X.C, Hu, Q.Y, Wei, T.B, Gan, J.H, Chen, H. | | 登録日 | 2016-08-03 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa
PLoS Pathog., 13, 2017
|
|
4DNW
 
 | | Crystal structure of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.773 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNV
 
 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
5GPE
 
 | | Crystal structure of the transcription regulator PbrR691 from Ralstonia metallidurans CH34 in complex with Lead(II) | | 分子名称: | LEAD (II) ION, Transcriptional regulator, MerR-family | | 著者 | Huang, S.Q, Chen, W.Z, Wang, D, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | 登録日 | 2016-08-01 | | 公開日 | 2016-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Basis for the Selective Pb(II) Recognition of Metalloregulatory Protein PbrR691
Inorg Chem, 55, 2016
|
|
2KXJ
 
 | | Solution structure of UBX domain of human UBXD2 protein | | 分子名称: | UBX domain-containing protein 4 | | 著者 | Wu, Q, Huang, H, Zhang, J, Hu, Q, Wu, J, Shi, Y. | | 登録日 | 2010-05-06 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution strcture of UBX domain of human UBXD2 protein
To be Published
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | 著者 | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | 登録日 | 2015-10-15 | | 公開日 | 2015-11-18 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | 分子名称: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | 著者 | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | 登録日 | 2022-04-28 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
5KNK
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii with catalytic residue substitution (E127A) | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | 著者 | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | 登録日 | 2016-06-28 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6WGN
 
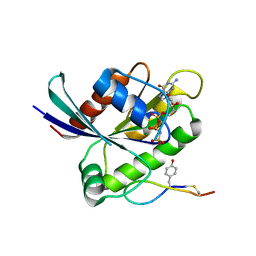 | | Crystal structure of K-Ras(G12D) GppNHp bound to cyclic peptide ligand KD2 | | 分子名称: | Cyclic Peptide KD2, GTPase KRas, MAGNESIUM ION, ... | | 著者 | Zhang, Z, Gao, R, Hu, Q, Peacock, H, Peacock, D.M, Shokat, K.M, Suga, H. | | 登録日 | 2020-04-06 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Acs Cent.Sci., 6, 2020
|
|
8K6C
 
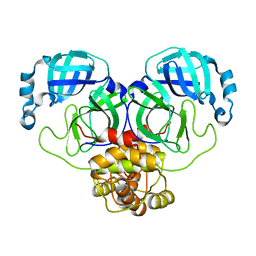 | |
8K68
 
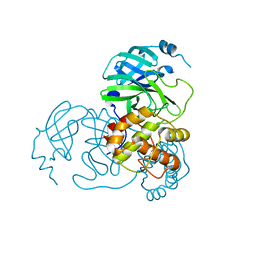 | |
8K6D
 
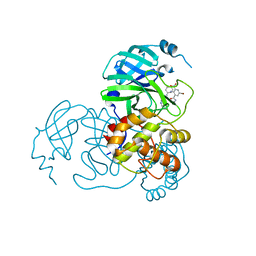 | |
8K6A
 
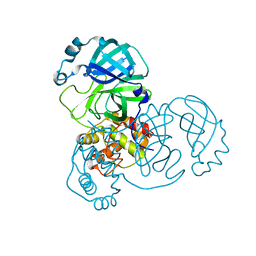 | |
8K6B
 
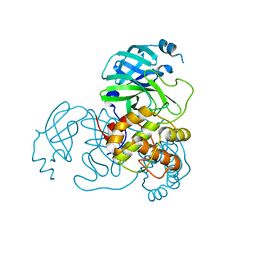 | |
8K67
 
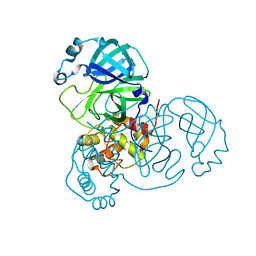 | |
8XL2
 
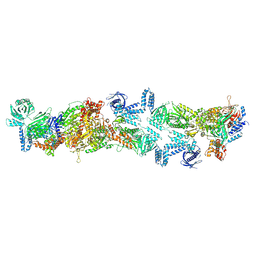 | | Human acetyl-CoA carboxylase 1 filament in complex with acetyl-CoA (ACC1-inact) | | 分子名称: | ACETYL COENZYME *A, Acetyl-CoA carboxylase 1 | | 著者 | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | 登録日 | 2023-12-25 | | 公開日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XL1
 
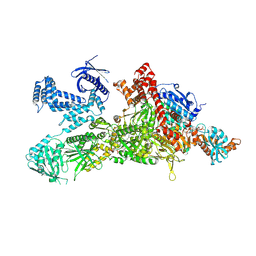 | | Core region of the human acetyl-CoA carboxylase 1 filament in complex with acetyl-CoA (ACC1-inact) | | 分子名称: | ACETYL COENZYME *A, Acetyl-CoA carboxylase 1 | | 著者 | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | 登録日 | 2023-12-25 | | 公開日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
