7Y6C
 
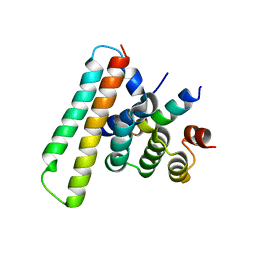 | | Crystal structure of the EscE/EsaG/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
7Y6B
 
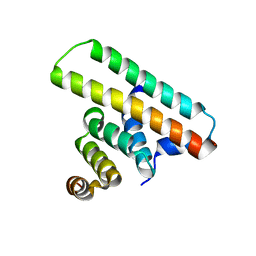 | | Crystal structure of the EscE/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
4M0W
 
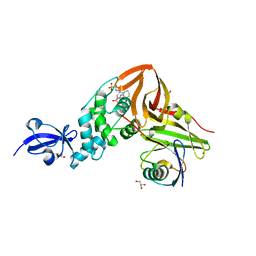 | | Crystal Structure of SARS-CoV papain-like protease C112S mutant in complex with ubiquitin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Replicase polyprotein 1a, ... | | Authors: | Chou, C.-Y, Chen, H.-Y, Lai, H.-Y, Cheng, S.-C, Chou, Y.-W. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3OOS
 
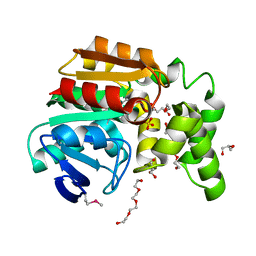 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
7X8X
 
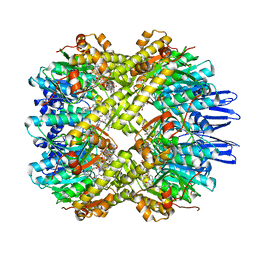 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
3OOP
 
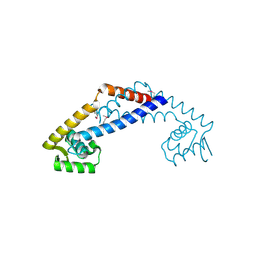 | | The structure of a protein with unknown function from Listeria innocua Clip11262 | | Descriptor: | Lin2960 protein | | Authors: | Fan, Y, Li, H, Zhou, Y, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of a protein with unknown function from Listeria innocua Clip11262
To be Published
|
|
6INW
 
 | | A Pericyclic Reaction enzyme | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Feng, Y, Chang, M, Wang, H, Liu, Z, Zhou, Y. | | Deposit date: | 2018-10-28 | | Release date: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
3O0J
 
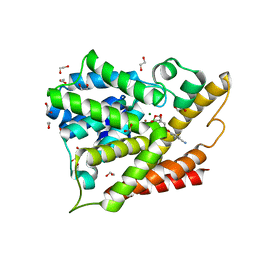 | | PDE4B In complex with ligand an2898 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)oxy]benzene-1,2-dicarbonitrile, MAGNESIUM ION, ... | | Authors: | Alley, M.R.K, Zhou, Y. | | Deposit date: | 2010-07-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Boron-based phosphodiesterase inhibitors show novel binding of boron to PDE4 bimetal center.
Febs Lett., 586, 2012
|
|
7X1U
 
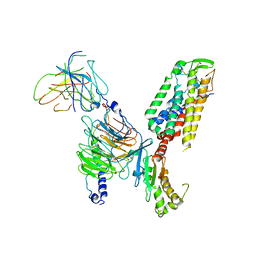 | | Structure of Thyrotropin-Releasing Hormone Receptor bound with an Endogenous Peptide Agonist TRH. | | Descriptor: | Endogenous Peptide Agonist TRH, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Zhang, H.H, Meng, X.Y, Li, Y.G, Zhou, Y.X, Ling, S.L, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into thyrotropin-releasing hormone receptor activation by an endogenous peptide agonist or its orally administered analogue.
Cell Res., 32, 2022
|
|
7XAT
 
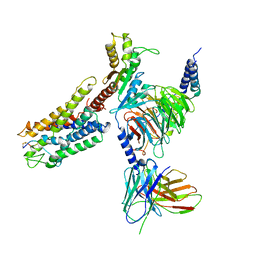 | | Structure of somatostatin receptor 2 bound with SST14. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAV
 
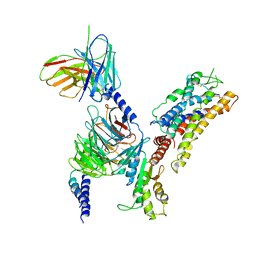 | | Structure of somatostatin receptor 2 bound with lanreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAU
 
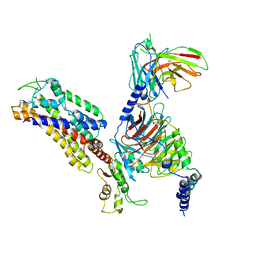 | | Structure of somatostatin receptor 2 bound with octreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7X1T
 
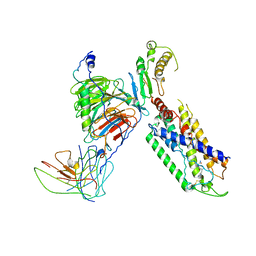 | | Structure of Thyrotropin-Releasing Hormone Receptor bound with Taltirelin. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ScFv16, ... | | Authors: | Yang, F, Zhang, H.H, Meng, X.Y, Li, Y.G, Zhou, Y.X, Ling, S.L, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into thyrotropin-releasing hormone receptor activation by an endogenous peptide agonist or its orally administered analogue.
Cell Res., 32, 2022
|
|
3BV9
 
 | | Structure of Thrombin Bound to the Inhibitor FM19 | | Descriptor: | FM19 inhibitor, GLYCEROL, IODIDE ION, ... | | Authors: | Nieman, M.T, Burke, F, Warnock, M, Zhou, Y, Sweigert, J, Chen, A, Ricketts, D, Lucchesi, B.R, Chen, Z, Di Cera, E, Hilfinger, J, Mosberg, H.I, Schmaier, A.H. | | Deposit date: | 2008-01-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thrombostatin FM compounds: direct thrombin inhibitors - mechanism of action in vitro and in vivo.
J.Thromb.Haemost., 6, 2008
|
|
6L15
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
5AX8
 
 | | Recombinant expression, purification and preliminary crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, mitochondrial | | Authors: | Jiang, X, Wang, J, Chang, H, Zhou, Y. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Recombinant expression, purification and crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase
Biosci Trends, 10, 2016
|
|
6L12
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L16
 
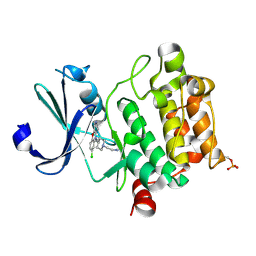 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-[4-[2-(7-chloranylpyrido[3,4-b][1,4]benzoxazin-5-yl)ethyl]piperidin-1-yl]ethanamine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L17
 
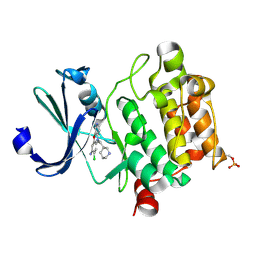 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L11
 
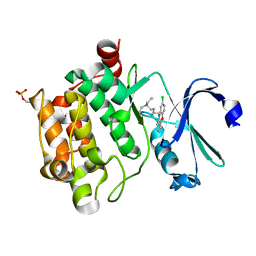 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 5-(2-chloranylphenoxazin-10-yl)-~{N},~{N}-diethyl-pentan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6K2U
 
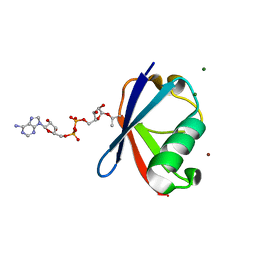 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
3RXJ
 
 | | Crystal structure of Trypsin complexed with 4-guanidinobenzoic acid | | Descriptor: | 4-carbamimidamidobenzoic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXU
 
 | | Crystal structure of Trypsin complexed with benzamide (F05 and A06, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXO
 
 | | Crystal structure of Trypsin complexed with (3-pyrrol-1-ylphenyl)methanamine | | Descriptor: | 1-[3-(1H-pyrrol-1-yl)phenyl]methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXI
 
 | | Crystal structure of Trypsin complexed with 2-(1H-indol-3-yl)ethanamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
