4ZVR
 
 | |
4ZVQ
 
 | |
4ZVP
 
 | |
4ZVO
 
 | |
1U3O
 
 | | Solution structure of rat Kalirin N-terminal SH3 domain | | Descriptor: | Huntingtin-associated protein-interacting protein | | Authors: | Schiller, M.R, Chakrabarti, K, King, G.F, Schiller, N.I, Eipper, B.A, Maciejewski, M.W. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regulation of RhoGEF Activity by Intramolecular and Intermolecular SH3 Domain Interactions.
J.Biol.Chem., 281, 2006
|
|
8SDX
 
 | | ATAD2B bromodomain in complex with histone H4 acetylated at lysine 5 with Serine 1 mutation to Cysteine | | Descriptor: | ATPase family AAA domain-containing protein 2B, SULFATE ION, histone H4S1CK5ac | | Authors: | Phillips, M, Montgomery, C, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-04-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
5JBM
 
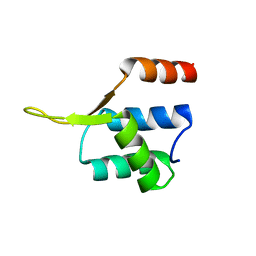 | | Crystal structgure of Cac1 C-terminus | | Descriptor: | Chromatin assembly factor 1 subunit p90 | | Authors: | Churchill, M.E.A, Liu, W, Zhou, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Cac1 subunit of histone chaperone CAF-1 organizes CAF-1-H3/H4 architecture and tetramerizes histones.
Elife, 5, 2016
|
|
8UHL
 
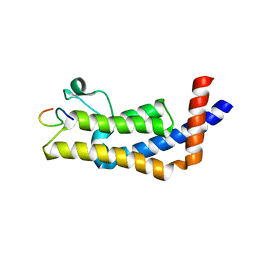 | | ATAD2B bromodomain in complex with histone H4 acetylated at lysine 12 | | Descriptor: | ATPase family AAA domain-containing protein 2B, Histone H4 | | Authors: | Phillips, M, Montgomery, C, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
2QRZ
 
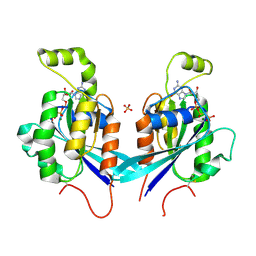 | | Cdc42 bound to GMP-PCP: Induced Fit by Effector is Required | | Descriptor: | Cell division control protein 42 homolog precursor, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Phillips, M.J, Calero, G, Chan, B, Cerione, R.A. | | Deposit date: | 2007-07-30 | | Release date: | 2008-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effector Proteins Exert an Important Influence on the Signaling-active State of the Small GTPase Cdc42.
J.Biol.Chem., 283, 2008
|
|
5BOO
 
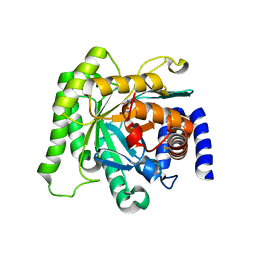 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Phillips, M, Deng, X, Tomchick, D. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
6CBQ
 
 | | Crystal structure of QscR bound to agonist S3 | | Descriptor: | (2S)-2-hexyl-N-[(3S)-2-oxooxolan-3-yl]decanamide, LuxR family transcriptional regulator | | Authors: | Churchill, M.E.A, Wysoczynski-Horita, C.L. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of agonism and antagonism of the Pseudomonas aeruginosa quorum sensing regulator QscR with non-native ligands.
Mol. Microbiol., 108, 2018
|
|
6CC0
 
 | |
5DEL
 
 | |
1FRG
 
 | |
3NM9
 
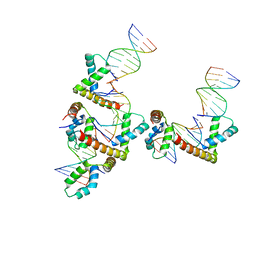 | | HMGD(M13A)-DNA complex | | Descriptor: | DNA 5'-D(*G*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3', High mobility group protein D | | Authors: | Churchill, M.E.A, Klass, J, Zoetewey, D.L. | | Deposit date: | 2010-06-22 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of HMGD-DNA complexes reveals influence of intercalation on sequence selectivity and DNA bending.
J.Mol.Biol., 403, 2010
|
|
3SZT
 
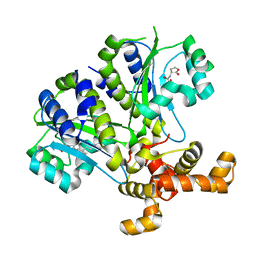 | | Quorum Sensing Control Repressor, QscR, Bound to N-3-oxo-dodecanoyl-L-Homoserine Lactone | | Descriptor: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, Quorum-sensing control repressor, SODIUM ION | | Authors: | Churchill, M.E.A, Lintz, M.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5K0U
 
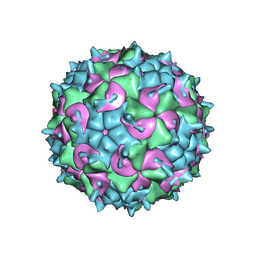 | | CryoEM structure of the full virion of a human rhinovirus C | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Hill, M.G, Klose, T, Chen, Z, Watters, K.E, Jiang, W, Palmenberg, A.C, Rossmann, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JZG
 
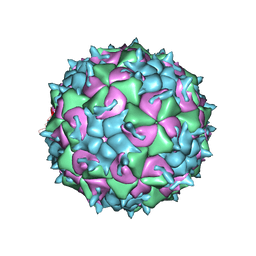 | | CryoEM structure of the native empty particle of a human rhinovirus C | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Liu, Y, Hill, M.G, Klose, T, Chen, Z, Watters, K.E, Jiang, W, Palmenberg, A.C, Rossmann, M.G. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6ZDF
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycoprotein endo-alpha-1,2-mannosidase | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZJ6
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with cyclohexylmethyl-Glc-1,3-isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ACETATE ION, ... | | Authors: | Thompson, A.J, Sobala, L.F, Fernandes, P.Z, Hakki, Z, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZFA
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with GlcIFG, alpha-1,2-mannobiose and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, 6-tungstotellurate(VI), ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZFN
 
 | | Structure of an inactive E404Q variant of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with 1-methyl alpha-1,2-mannobiose | | Descriptor: | Glycoprotein endo-alpha-1,2-mannosidase, SULFATE ION, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDC
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with nickel | | Descriptor: | Glycoprotein endo-alpha-1,2-mannosidase, NICKEL (II) ION | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZFQ
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with bis-tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoprotein endo-alpha-1,2-mannosidase | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDL
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with GlcIFG and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, 6-tungstotellurate(VI), ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
