3K79
 
 | | C38A, C52V Cysteine-Free Variant of Rop (Rom) | | Descriptor: | Regulatory protein rop | | Authors: | Hari, S.B, Magliery, T.J. | | Deposit date: | 2009-10-12 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Cysteine-free Rop: a four-helix bundle core mutant has wild-type stability and structure but dramatically different unfolding kinetics.
Protein Sci., 19, 2010
|
|
4I5H
 
 | |
6BQC
 
 | | Cyclopropane fatty acid synthase from E. coli | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CARBONATE ION, Cyclopropane-fatty-acyl-phospholipid synthase, ... | | Authors: | Hari, S.B, Grant, R.A, Sauer, R.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Structural and Functional Analysis of E. coli Cyclopropane Fatty Acid Synthase.
Structure, 26, 2018
|
|
4N4S
 
 | | A Double Mutant Rat Erk2 in Complex With a Pyrazolo[3,4-d]pyrimidine Inhibitor | | Descriptor: | 3-[2-(benzyloxy)-8-methylquinolin-6-yl]-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Mitogen-activated protein kinase 1 | | Authors: | Hari, S.B, Maly, D.J, Merritt, E.A. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-Selective ATP-Competitive Inhibitors Control Regulatory Interactions and Noncatalytic Functions of Mitogen-Activated Protein Kinases.
Chem.Biol., 21, 2014
|
|
7NVR
 
 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
1K4Y
 
 | | Crystal Structure of Rabbit Liver Carboxylesterase in Complex with 4-piperidino-piperidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-PIPERIDINO-PIPERIDINE, LIVER CARBOXYLESTERASE, ... | | Authors: | Bencharit, S, Morton, C.L, Howard-Williams, E.L, Danks, M.K, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2001-10-09 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into CPT-11 activation by mammalian carboxylesterases.
Nat.Struct.Biol., 9, 2002
|
|
3RM3
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
8RAO
 
 | | Structure of Sen1-ADP.BeF3-RNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Helicase SEN1, ... | | Authors: | Rengachari, S, Lidscreiber, M, Cramer, P. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-30 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of polyadenylation-independent RNA polymerase II termination.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8RAN
 
 | |
3RLI
 
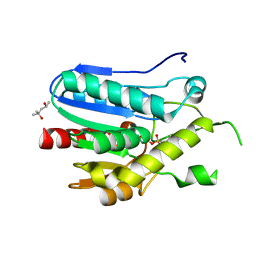 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 in complex with PMSF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase, phenylmethanesulfonic acid | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-19 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
2GEE
 
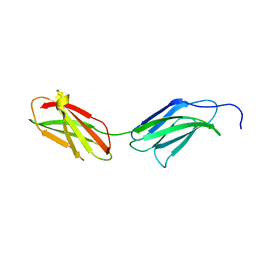 | | Crystal Structure of Human Type III Fibronectin Extradomain B and Domain 8 | | Descriptor: | hypothetical protein | | Authors: | Bencharit, S, Cui, C.B, Siddiqui, A, Howard-Williams, E.L, Aukhil, I. | | Deposit date: | 2006-03-19 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into fibronectin type III domain-mediated signaling.
J.Mol.Biol., 367, 2007
|
|
1MX9
 
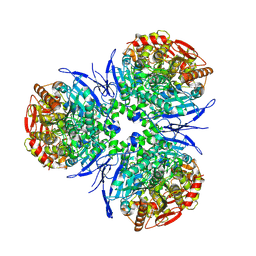 | | Crystal Structure of Human Liver Carboxylesterase in complexed with naloxone methiodide, a heroin analogue | | Descriptor: | (5A,17R)-4,5-EPOXY-3,14-DIHYDROXY-17-METHYL-6-OXO-17-(2-PROPENYL)-MORPHINANIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, liver Carboxylesterase I | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
1MX5
 
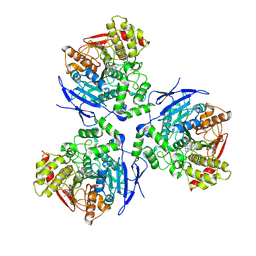 | | Crystal Structure of Human Liver Carboxylesterase in complexed with homatropine, a cocaine analogue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, HOMOTROPINE, ... | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
4KE8
 
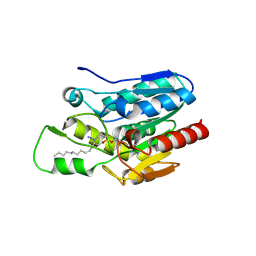 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with monopalmitoyl glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, tetradecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
8RAM
 
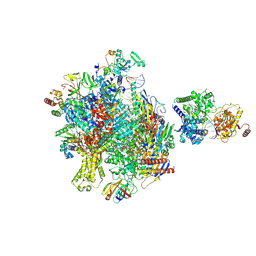 | | Structure of Sen1 bound RNA Polymerase II pre-termination complex | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Rengachari, S, Lidscreiber, M, Cramer, P. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-30 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of polyadenylation-independent RNA polymerase II termination.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8RAP
 
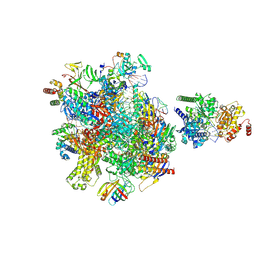 | | Structure of Sen1-ADP.BeF3 bound RNA Polymerase II pre-termination complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Rengachari, S, Lidscreiber, M, Cramer, P. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-30 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of polyadenylation-independent RNA polymerase II termination.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5OEN
 
 | | Crystal Structure of STAT2 in complex with IRF9 | | Descriptor: | Interferon regulatory factor 9, Signal transducer and activator of transcription | | Authors: | Rengachari, S, Panne, D. | | Deposit date: | 2017-07-09 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.919 Å) | | Cite: | Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ZWC
 
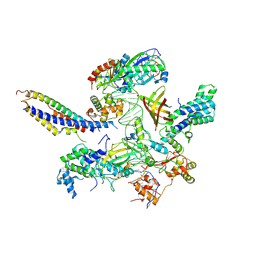 | | Structure of SNAPc:TBP-TFIIA-TFIIB sub-complex bound to U5 snRNA promoter | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8RWB
 
 | | Crystal structure of ULBP6 in complex with a blocking antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bharill, S, Chen, I, Ivic, N, Bahrami Dizicheh, Z, Wu, Y, Doamekpor, S, Koenig, P, Fuh, G. | | Deposit date: | 2024-02-02 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Crystal structure of ULBP6 in complex with a blocking antibody
To Be Published
|
|
5OEM
 
 | |
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4KE6
 
 | | Crystal structure D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in complex with 1-rac-lauroyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE9
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-stearyol glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, hexadecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
1MX1
 
 | | Crystal Structure of Human Liver Carboxylesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Bencharit, S, Morton, C.L, Hyatt, J.L, Kuhn, P, Danks, M.K, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Carboxylesterase 1 Complexed with the Alzheimer's
Drug Tacrine: From Binding Promiscuity to Selective Inhibition
CHEM.BIOL., 10, 2003
|
|
7ZWD
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
