4IAQ
 
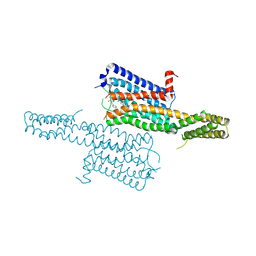 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
7U0Z
 
 | | High-resolution map of tau filament from progressive supranuclear palsy (PSP) case 1 | | Descriptor: | Isoform Tau-E of Microtubule-associated protein tau | | Authors: | FItzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
2GM8
 
 | | TenA Homolog/Thi-4 Thiaminase complexed with product 4-amino-5-hydroxymethyl-2-methylpyrimidine | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, tenA homolog/Thi-4 Thiaminase | | Authors: | Sawaya, M.R, Chan, S, Han, G.W, Perry, L.J, Pashkov, I. | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a TenA Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum
To be Published
|
|
2GM7
 
 | | TenA Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sawaya, M.R, Chan, S, Han, G.W, Perry, L.J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Ten A Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum
To be Published
|
|
4DJH
 
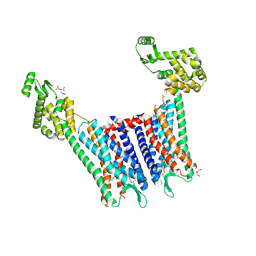 | | Structure of the human kappa opioid receptor in complex with JDTic | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (3R)-7-hydroxy-N-{(2S)-1-[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]-3-methylbutan-2-yl}-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CITRIC ACID, ... | | Authors: | Wu, H, Wacker, D, Katritch, V, Mileni, M, Han, G.W, Vardy, E, Liu, W, Thompson, A.A, Huang, X.P, Carroll, F.I, Mascarella, S.W, Westkaemper, R.B, Mosier, P.D, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-02-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | tructure of the human kappa-opioid receptor in complex with JDTic
Nature, 485, 2012
|
|
4OR2
 
 | | Human class C G protein-coupled metabotropic glutamate receptor 1 in complex with a negative allosteric modulator | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{4-[6-(propan-2-ylamino)pyrimidin-4-yl]-1,3-thiazol-2-yl}benzamide, CHOLESTEROL, ... | | Authors: | Wu, H, Wang, C, Gregory, K.J, Han, G.W, Cho, H.P, Xia, Y, Niswender, C.M, Katritch, V, Cherezov, V, Conn, P.J, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator
Science, 344, 2014
|
|
1Y2M
 
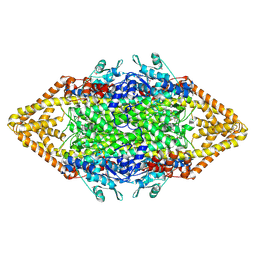 | | Crystal structure of phenylalanine ammonia-lyase from yeast Rhododporidium toruloides | | Descriptor: | Phenylalanine ammonia-lyase | | Authors: | Wang, L, Gamez, A, Sarkissian, C.N, Straub, M, Patch, M.G, Han, G.W, Scriver, C.R, Stevens, R.C. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based chemical modification strategy for enzyme replacement treatment of phenylketonuria.
Mol.Genet.Metab., 86, 2005
|
|
4N4W
 
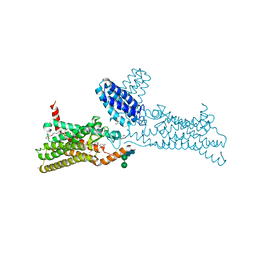 | | Structure of the human smoothened receptor in complex with SANT-1. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (E)-N-(4-benzylpiperazin-1-yl)-1-(3,5-dimethyl-1-phenyl-1H-pyrazol-4-yl)methanimine, Cytochrome b(562),Smoothened homolog, ... | | Authors: | Wang, C, Wu, H, Han, G.W, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
4QIN
 
 | | Structure of the human smoothened receptor in complex with SAG1.5 | | Descriptor: | 3-chloro-4,7-difluoro-N-[trans-4-(methylamino)cyclohexyl]-N-[3-(pyridin-4-yl)benzyl]-1-benzothiophene-2-carboxamide, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
4QIM
 
 | | Structure of the human smoothened receptor in complex with ANTA XV | | Descriptor: | 2-{6-[4-(4-benzylphthalazin-1-yl)piperazin-1-yl]pyridin-3-yl}propan-2-ol, Smoothened homolog/Soluble cytochrome b562 chimeric protein, ZINC ION | | Authors: | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
9EOQ
 
 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
7K15
 
 | | Crystal structure of the Human Leukotriene B4 Receptor 1 in Complex with Selective Antagonist MK-D-046 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Michaelian, N, Han, G.W, Cherezov, V. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights on ligand recognition at the human leukotriene B4 receptor 1.
Nat Commun, 12, 2021
|
|
6LVB
 
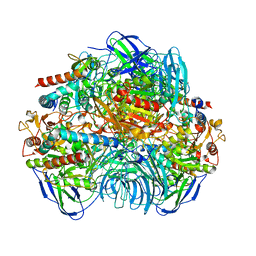 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7JNI
 
 | | Crystal structure of the angiotensin II type 2 receptoror (AT2R) in complex with EMA401 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FORMIC ACID, HEXANE-1,6-DIOL, ... | | Authors: | Cherezov, V, Shaye, H, Han, G.W. | | Deposit date: | 2020-08-04 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of the angiotensin II type 2 receptor AT 2 R is a novel therapeutic strategy for glioblastoma.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6Q2R
 
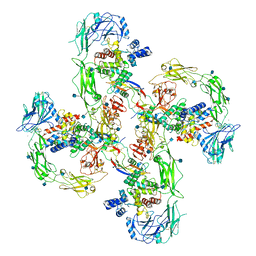 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex in the tetrameric form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2J
 
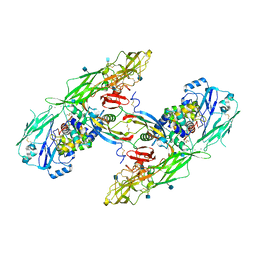 | | Cryo-EM structure of extracellular dimeric complex of RET/GFRAL/GDF15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-like, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2S
 
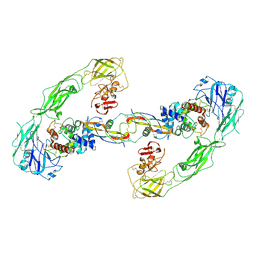 | | Cryo-EM structure of RET/GFRa3/ARTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-3, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2O
 
 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2N
 
 | | Cryo-EM structure of RET/GFRa1/GDNF extracellular complex | | Descriptor: | CALCIUM ION, GDNF family receptor alpha-1, Glial cell line-derived neurotrophic factor, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
1PJO
 
 | | Crystal Structure of an RNA/DNA hybrid of HIV-1 PPT | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', MAGNESIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An Unusual Sugar Conformation in the Structure of an RNA/DNA
Decamer of the Polypurine Tract May Affect Recognition by RNase H
J.Mol.Biol., 334, 2003
|
|
1PJG
 
 | | RNA/DNA Hybrid Decamer of CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', CALCIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H.
J.Mol.Biol., 334, 2003
|
|
6LI1
 
 | | Crystal structure of GPR52 ligand free form with flavodoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Flavodoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
4MMP
 
 | |
6LVE
 
 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
