3JYN
 
 | | Crystal structures of Pseudomonas syringae pv. Tomato DC3000 quinone oxidoreductase complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Quinone oxidoreductase | | Authors: | Pan, X, Zhang, H, Gao, Y, Li, M, Chang, W. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of Pseudomonas syringae pv. tomato DC3000 quinone oxidoreductase and its complex with NADPH
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3JYL
 
 | | Crystal structures of Pseudomonas syringae pv. Tomato DC3000 quinone oxidoreductase | | Descriptor: | Quinone oxidoreductase | | Authors: | Pan, X, Zhang, H, Gao, Y, Li, M, Chang, W. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Pseudomonas syringae pv. tomato DC3000 quinone oxidoreductase and its complex with NADPH
Biochem.Biophys.Res.Commun., 390, 2009
|
|
4NQ0
 
 | | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones | | Descriptor: | HAT1-interacting factor 1 | | Authors: | Liu, H, Zhang, M, He, W, Zhu, Z, Teng, M, Gao, Y, Niu, L. | | Deposit date: | 2013-11-23 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones
Biochem.J., 462, 2014
|
|
4OQT
 
 | | LINGO-1/Li81 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Li81 Fab, ... | | Authors: | Pepinsky, R.B, Arndt, J.W, Quan, C, Gao, Y, Quintero-Monzon, O, Lee, X, Mi, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the LINGO-1-Anti-LINGO-1 Li81 Antibody Complex Provides Insights into the Biology of LINGO-1 and the Mechanism of Action of the Antibody Therapy.
J.Pharmacol.Exp.Ther., 350, 2014
|
|
7XR2
 
 | |
8AKO
 
 | | Structure of EspB-EspK complex: the non-identical twin of the PE-PPE-EspG secretion mechanism. | | Descriptor: | ESX-1 secretion-associated protein EspB, ESX-1 secretion-associated protein EspK | | Authors: | Gijsbers, A, Eymery, M, Menart, I, Vinciauskaite, V, Gao, Y, Siliqi, D, Peters, P, Mccarthy, A, Ravelli, R.B.G. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in Mycobacterium tuberculosis.
J.Biol.Chem., 299, 2022
|
|
6IQJ
 
 | |
7EGQ
 
 | | Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L.M, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading.
Cell, 184, 2021
|
|
4KLY
 
 | | Crystal structure of a blue-light absorbing proteorhodopsin mutant D97N from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KNF
 
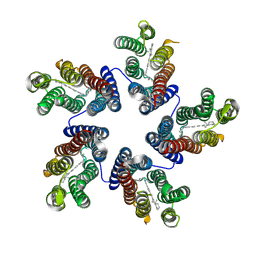 | | Crystal structure of a blue-light absorbing proteorhodopsin double-mutant D97N/Q105L from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
6IQI
 
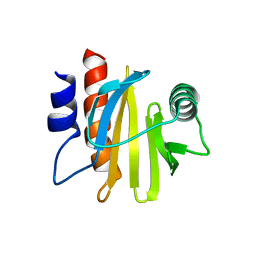 | | crystal structure of Arabidopsis thaliana Profilin 2 | | Descriptor: | Profilin-2 | | Authors: | Qiao, Z, Gao, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and computational examination of theArabidopsisprofilin-Poly-P complex reveals mechanistic details in profilin-regulated actin assembly.
J.Biol.Chem., 294, 2019
|
|
7EIZ
 
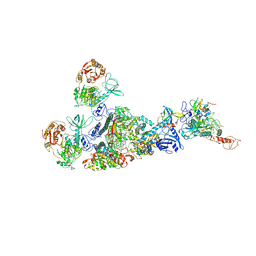 | | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-22 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading
Cell, 184, 2021
|
|
8GWO
 
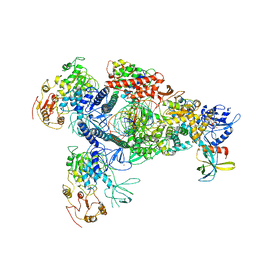 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWG
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWI
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWN
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWK
 
 | | SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
3UNW
 
 | | Crystal Structure of Human GAC in Complex with Glutamate | | Descriptor: | GLUTAMIC ACID, Glutaminase kidney isoform, mitochondrial | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wie, W, Hurov, J. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
3UO9
 
 | | Crystal Structure of Human GAC in Complex with Glutamate and BPTES | | Descriptor: | GLYCEROL, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wei, W, Hurov, J.B. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
4GBV
 
 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase Mutant A54L with 1,2-ethanediol as Cryo-protectant | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Honzatko, R.B, Gao, Y. | | Deposit date: | 2012-07-28 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Water Structure of the Central Hydrophobic Cavity of Mammalian Fructose-1,6-bisphosphatase: a Potential Thermodynamic Determinant of Allowed Quaternary States
To be published
|
|
7TIL
 
 | | CryoEM structure of JetD from Pseudomonas aeruginosa | | Descriptor: | JetD | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-01-13 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
4HG3
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-06 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H5U
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 | | Descriptor: | CACODYLATE ION, GLYCEROL, Probable hydrolase NIT2 | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
