6DT0
 
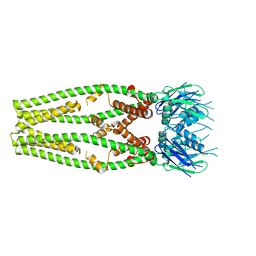 | | Cryo-EM structure of a mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter | | Authors: | Yoo, J, Wu, M, Yin, Y, Herzik, M.A.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a mitochondrial calcium uniporter.
Science, 361, 2018
|
|
5AN8
 
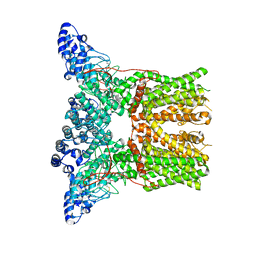 | | Cryo-electron microscopy structure of rabbit TRPV2 ion channel | | Descriptor: | TRPV2 | | Authors: | Zubcevic, L, Herzik, M.A.J, Chung, B.C, Lander, G.C, Lee, S.Y. | | Deposit date: | 2015-09-04 | | Release date: | 2015-12-23 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-Electron Microscopy of the Trpv2 Ion Channel
Nat.Struct.Mol.Biol., 23, 2016
|
|
6V21
 
 | |
8DPN
 
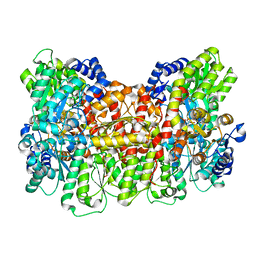 | | CryoEM structure of Azotobacter vinelandii nitrogenase MoFeP during catalytic N2 reduction | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-07-15 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
6V20
 
 | |
8DK2
 
 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetABC in an unclamped state trapped in ATP dependent dimeric form | | Descriptor: | DNA (26-MER), JetA, JetB, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
7UT6
 
 | | C1 symmetric cryoEM structure of Azotobacter vinelandii MoFeP under non-turnover conditions | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.91 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT8
 
 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (1:1 FeP:MoFeP, ATP-bound) during catalytic N2 reduction | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | Authors: | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT9
 
 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (1:1 FeP:MoFeP, ADP/ATP-bound) during catalytic N2 reduction | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
8DK3
 
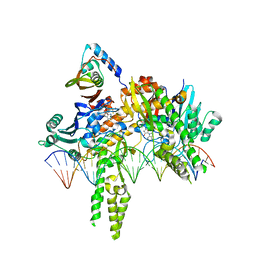 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetC ATPase domain bound to DNA and cWHD domain of JetA | | Descriptor: | DNA (26-MER), JetA, JetC, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
8DK1
 
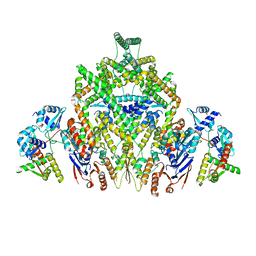 | | CryoEM structure of JetABC (head construct) from Pseudomonas aeruginosa PA14 | | Descriptor: | JetA, JetB, JetC | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
6MHS
 
 | | Structure of the human TRPV3 channel in a putative sensitized conformation | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
6MHX
 
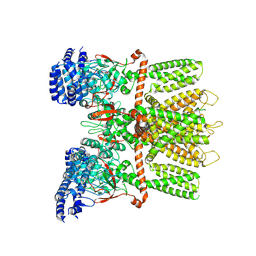 | | Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (2) | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
6MHW
 
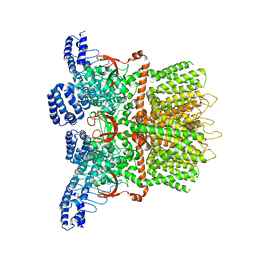 | | Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (1) | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
6MHV
 
 | | Structure of human TRPV3 in the presence of 2-APB in C4 symmetry | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
7UTA
 
 | | CryoEM structure of Azotobacter vinelandii nitrogenase complex (2:1 FeP:MoFeP) inhibited by BeFx during catalytic N2 reduction | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM, ... | | Authors: | Rutledge, H.L, Cook, B.D, Nguyen, H.P.M, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT7
 
 | | C2 symmetric cryoEM structure of Azotobacter vinelandii MoFeP under non-turnover conditions | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Cook, B.D, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.91 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7TIL
 
 | | CryoEM structure of JetD from Pseudomonas aeruginosa | | Descriptor: | JetD | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-01-13 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
6MHO
 
 | | Structure of the human TRPV3 channel in the apo conformation | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
5W3S
 
 | | Cryo-electron microscopy structure of a TRPML3 ion channel | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Mucolipin-3 isoform 1, ... | | Authors: | Hirschi, M, Herzik, M.A, Wie, J, Suo, Y, Borschel, W.F, Ren, D, Lander, G.C, Lee, S.Y. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3.
Nature, 550, 2017
|
|
