2I3I
 
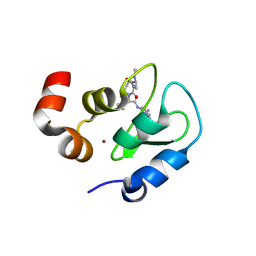 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | (3R,6R,9AR)-2,2-DIMETHYL-6-[(N-METHYL-L-ALANYL)AMINO]-N-(3-METHYL-1-PHENYL-1H-PYRAZOL-5-YL)-5-OXO-2,3,5,6,9,9A-HEXAHYDRO[1,3]THIAZOLO[3,2-A]AZEPINE-3-CARBOXAMIDE, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fairbrother, W.J, Franklin, M.C. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs.
Acs Chem.Biol., 1, 2006
|
|
8U4N
 
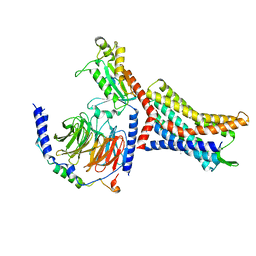 | | Structure of Apo CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 2024
|
|
8U4T
 
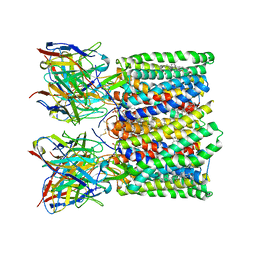 | |
8U4R
 
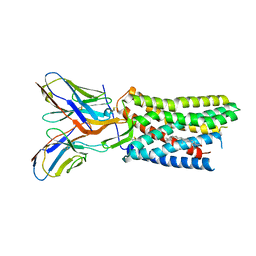 | |
8U4P
 
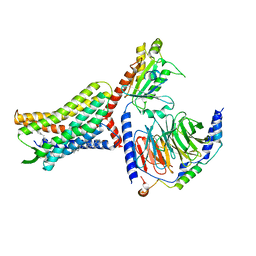 | | Structure of AMD3100-bound CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 2024
|
|
8U4S
 
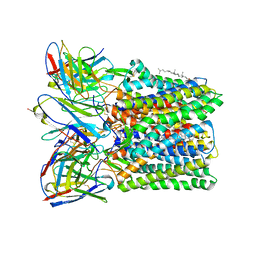 | |
8U4O
 
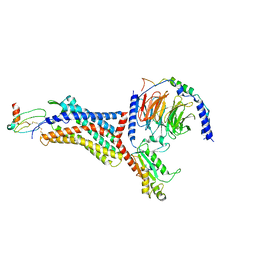 | | Structure of CXCL12-bound CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 2024
|
|
8U4Q
 
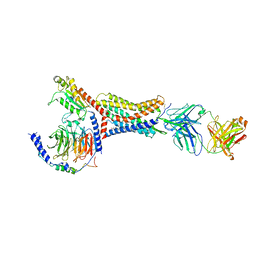 | |
8ESA
 
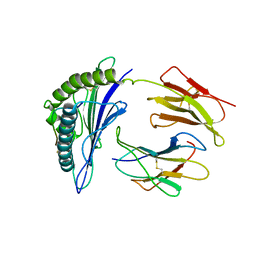 | |
8ES7
 
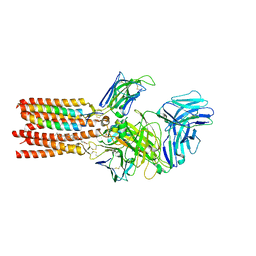 | | CryoEM structure of PN45545 TCR-CD3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM.
Nat Commun, 14, 2023
|
|
8ESB
 
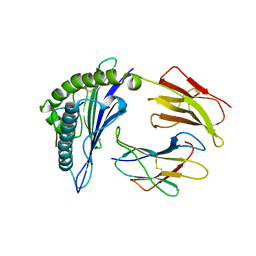 | |
8ES9
 
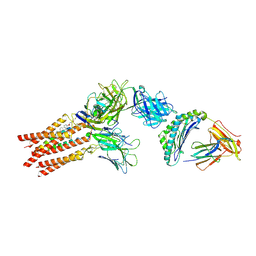 | | CryoEM structure of PN45428 TCR-CD3 in complex with HLA-A2 MAGEA4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM.
Nat Commun, 14, 2023
|
|
8ES8
 
 | |
8FJB
 
 | |
8FJA
 
 | |
1HT2
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
8D74
 
 | |
8D7R
 
 | |
8D7E
 
 | |
8D6A
 
 | |
8D7H
 
 | | Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cardiotrophin-like cytokine factor 1, ... | | Authors: | Zhou, Y, Franklin, M.C. | | Deposit date: | 2022-06-07 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly of gp130 family cytokine signaling complexes.
Sci Adv, 9, 2023
|
|
4NBQ
 
 | | Structure of the polynucleotide phosphorylase (CBU_0852) from Coxiella burnetii | | Descriptor: | Polyribonucleotide nucleotidyltransferase, SULFATE ION | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2013-10-23 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9138 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
7M42
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of neutralizing antibodies REGN10985 and REGN10989 | | Descriptor: | REGN10985 antibody Fab fragment heavy chain, REGN10985 antibody Fab fragment light chain, REGN10989 antibody Fab fragment heavy chain, ... | | Authors: | Zhou, Y, Romero Hernandez, A, Saotome, K, Franklin, M.C. | | Deposit date: | 2021-03-19 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies.
Cell, 184, 2021
|
|
7MXL
 
 | |
4WB8
 
 | | Crystal structure of human cAMP-dependent protein kinase A (catalytic alpha subunit), exon 1 deletion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
