4WRL
 
 | | Structure of the human CSF-1:CSF-1R complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage colony-stimulating factor 1, Macrophage colony-stimulating factor 1 receptor, ... | | Authors: | Felix, J, De Munck, S, Elegheert, J, Savvides, S.N. | | Deposit date: | 2014-10-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure and Assembly Mechanism of the Signaling Complex Mediated by Human CSF-1.
Structure, 23, 2015
|
|
4WRM
 
 | | Structure of the human CSF-1:CSF-1R complex | | Descriptor: | Macrophage colony-stimulating factor 1, Macrophage colony-stimulating factor 1 receptor | | Authors: | Felix, J, De Munck, S, Elegheert, J, Savvides, S.N. | | Deposit date: | 2014-10-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (6.853 Å) | | Cite: | Structure and Assembly Mechanism of the Signaling Complex Mediated by Human CSF-1.
Structure, 23, 2015
|
|
5D28
 
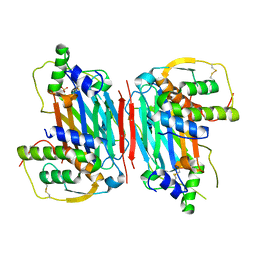 | |
7PQH
 
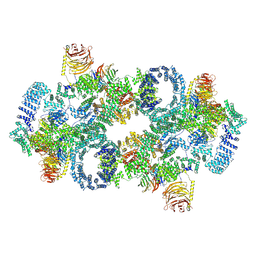 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5D22
 
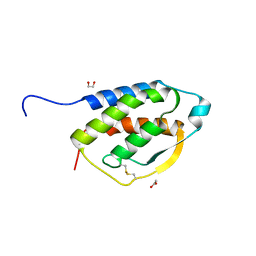 | |
6Y3X
 
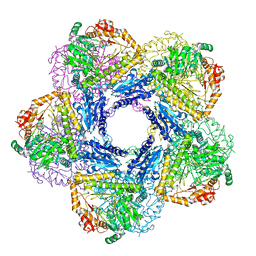 | |
6YN6
 
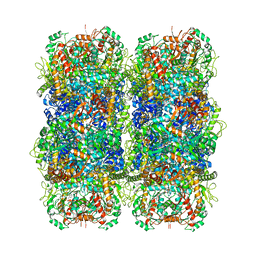 | | Inducible lysine decarboxylase LdcI stacks, pH 5.7 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Felix, J, Jessop, M, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
9G5I
 
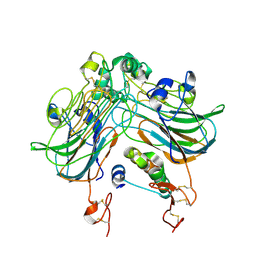 | | Cryo-EM structure of a 2:1 ALK:ALKAL2 complex obtained after re-processing of EMPIAR-10930 data | | Descriptor: | ALK and LTK ligand 2, ALK tyrosine kinase receptor | | Authors: | Felix, J, De Munck, S, Bazan, J.F, Savvides, S.N. | | Deposit date: | 2024-07-17 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Reanalysis of cryo-EM data reveals ALK-cytokine assemblies with both 2:1 and 2:2 stoichiometries.
Plos Biol., 23, 2025
|
|
6HWN
 
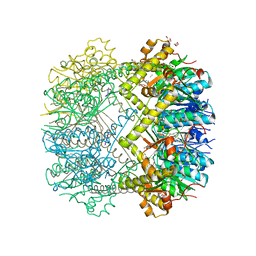 | | Structure of Thermus thermophilus ClpP in complex with a tripeptide. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, Unknown tripeptide | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
6HWM
 
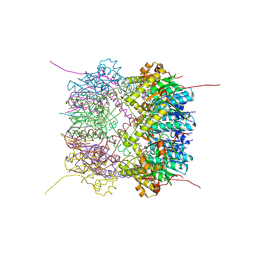 | | Structure of Thermus thermophilus ClpP in complex with bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
8REW
 
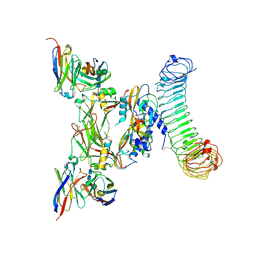 | | CryoEM structure of human GARP-lTGFbeta1 in complex with a Fab fragment derived from an activating antibody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta activator LRRC32, ... | | Authors: | Felix, J, Lambert, F, Marien, L, van der Woning, B, Savvides, S.N, Lucas, S. | | Deposit date: | 2023-12-12 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | TGF-beta1 activating antibodies as novel immunotherapeutics for graft-versus-host disease.
To Be Published
|
|
8A8V
 
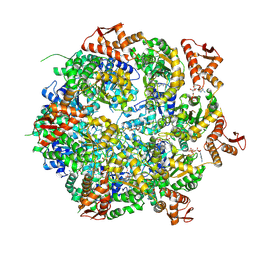 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8W
 
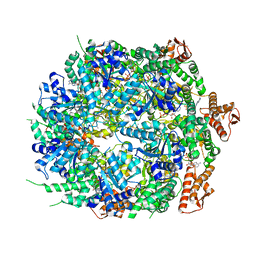 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8U
 
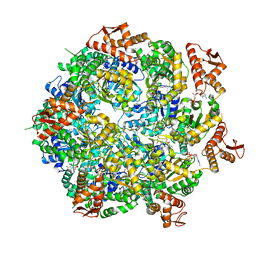 | | Mycobacterium tuberculosis ClpC1 hexamer structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8ODZ
 
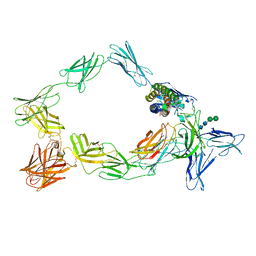 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, Interleukin-12 receptor subunit beta-2,Calmodulin-1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PB1
 
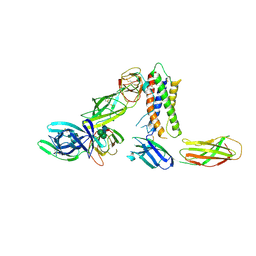 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1), obtained after local refinement. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, Interleukin-12 receptor subunit beta-2,Calmodulin-1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-06-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OE0
 
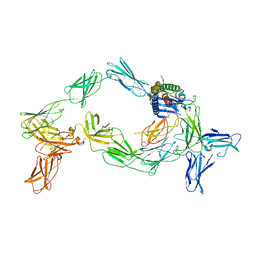 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 2). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8CR6
 
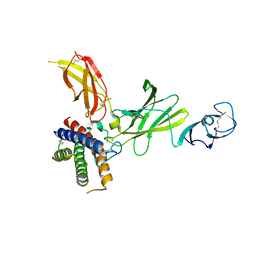 | | mouse Interleukin-12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 subunit alpha, Interleukin-12 subunit beta, ... | | Authors: | Merceron, R, Felix, J, Lambert, E, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8CR5
 
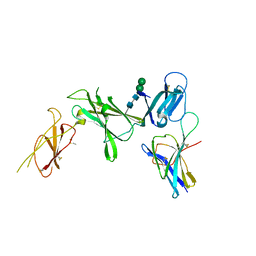 | | Murine Interleukin-12 receptor beta 1 domain 1 in complex with murine Interleukin-12 beta. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, Interleukin-12 subunit beta, ... | | Authors: | Merceron, R, Bloch, Y, Felix, J, Savvides, S.N. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6JJJ
 
 | |
6YN5
 
 | | Inducible lysine decarboxylase LdcI decamer, pH 7.0 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Jessop, M, Felix, J, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CCH
 
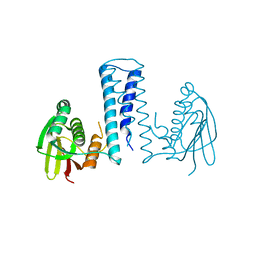 | | Acinetobacter baumannii histidine kinase AdeS | | Descriptor: | AdeS | | Authors: | Wen, Y, Felix, J. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Proteolysis and multimerization regulate signaling along the two-component regulatory system AdeRS.
Iscience, 24, 2021
|
|
6Q7M
 
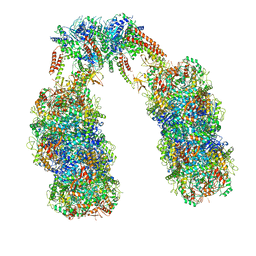 | | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RavA, Inducible lysine decarboxylase, ... | | Authors: | Arragain, B, Felix, J, Malet, H, Gutsche, I, Jessop, M. | | Deposit date: | 2018-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex.
Commun Biol, 3, 2020
|
|
6Q7L
 
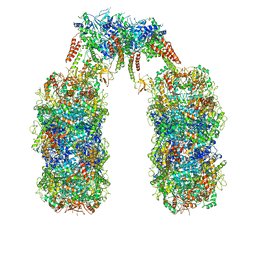 | | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RavA, Inducible lysine decarboxylase, ... | | Authors: | Arragain, B, Felix, J, Malet, H, Gutsche, I, Jessop, M. | | Deposit date: | 2018-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex.
Commun Biol, 3, 2020
|
|
6SZA
 
 | |
