1E4Y
 
 | |
6BZ7
 
 | | Thermus thermophilus 70S containing 16S G299A point mutation and near-cognate ASL Leucine in A site. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
8VJB
 
 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
4S3F
 
 | | IspG in complex with Inhibitor 8 (compound 1077) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, FE3-S4 CLUSTER, but-3-yn-1-yl trihydrogen diphosphate | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
1VTX
 
 | |
2Q4O
 
 | | Ensemble refinement of the crystal structure of a lysine decarboxylase-like protein from Arabidopsis thaliana gene At2g37210 | | Descriptor: | MAGNESIUM ION, SULFATE ION, Uncharacterized protein At2g37210/T2N18.3 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4Z
 
 | | Ensemble refinement of the protein crystal structure of an aspartoacylase from Rattus norvegicus | | Descriptor: | Aspartoacylase, SULFATE ION, ZINC ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
1W1X
 
 | | Structure of Neuraminidase from English duck subtype N6 complexed with 30 mM sialic acid (NANA, Neu5Ac), crystal soaked for 3 hours at 277 K. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2004-06-24 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Type a Influenza Virus Neuraminidase of the N6 Subtype Reveals the Existence of Two Separate Neu5Ac Binding Sites
To be Published
|
|
6C23
 
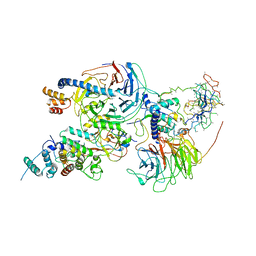 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
3JVA
 
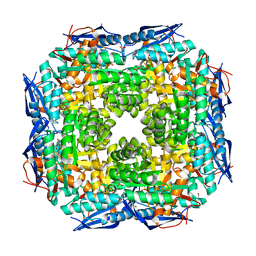 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 | | Descriptor: | Dipeptide Epimerase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Imker, H, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7UUQ
 
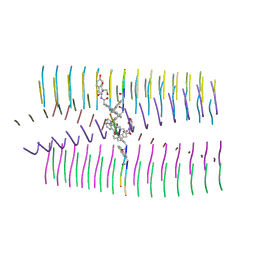 | |
5D1R
 
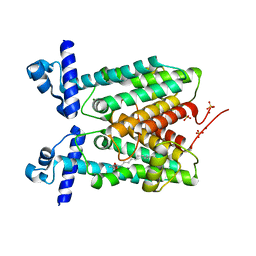 | | Crystal structure of Mycobacterium tuberculosis Rv1816 transcriptional regulator. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Rv1816 transcriptional regulator, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
1EFC
 
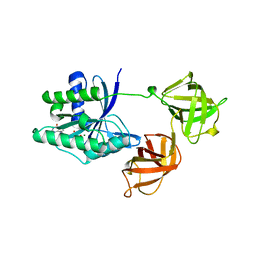 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
1MKD
 
 | | crystal structure of PDE4D catalytic domain and zardaverine complex | | Descriptor: | 6-(4-DIFLUOROMETHOXY-3-METHOXY-PHENYL)-2H-PYRIDAZIN-3-ONE, MAGNESIUM ION, Phosphodiesterase 4D, ... | | Authors: | Lee, M.E, Markowitz, J, Lee, J.-O, Lee, H. | | Deposit date: | 2002-08-29 | | Release date: | 2003-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of phosphodiesterase 4D and inhibitor complex
FEBS LETT., 530, 2002
|
|
7UY4
 
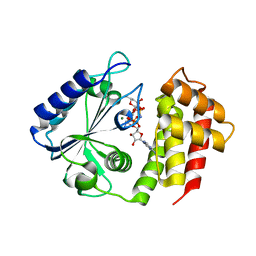 | | Aminoglycoside-modifying enzyme ANT-3,9 in complex with spectinomycin and AMP-PNP | | Descriptor: | Aminoglycoside (3'') (9) adenylyltransferase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Reeve, S.M, Lee, R.E, Das, N, Jayaraman, S. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Characterization of the spectinomycin deactivating enzyme ANT-3,9
To Be Published
|
|
3JYJ
 
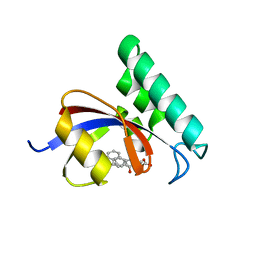 | | Structure-Based Design of Novel PIN1 Inhibitors (II) | | Descriptor: | (2R,4E)-2-[(naphthalen-2-ylcarbonyl)amino]-5-phenylpent-4-enoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2009-09-21 | | Release date: | 2010-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (II).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6BW5
 
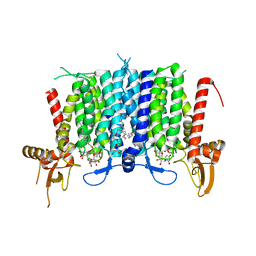 | | Human GPT (DPAGT1) in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8P5F
 
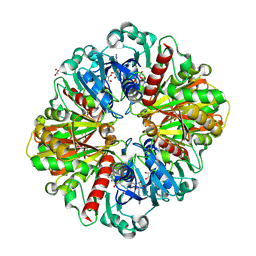 | | Human wild-type GAPDH,orthorhombic form | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Samygina, V.R, Muronetz, V.I, Schmalhausen, E.V. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S-nitrosylation and S-glutathionylation of GAPDH: Similarities, differences, and relationships.
Biochim Biophys Acta Gen Subj, 1867, 2023
|
|
7T0W
 
 | | Complex of GABA-A synaptic receptor with autoimmune antibody Fab115 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab115 Heavy Chain, IgG1, ... | | Authors: | Noviello, C.M, Hibbs, R.E, Kreye, J, Teng, J, Pruss, H. | | Deposit date: | 2021-11-30 | | Release date: | 2022-07-13 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanisms of GABA A receptor autoimmune encephalitis.
Cell, 185, 2022
|
|
8P5O
 
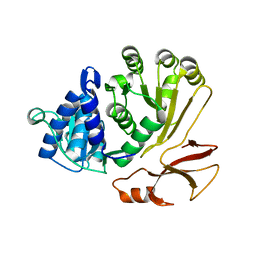 | | Proline activating adenylation domain of gramicidin S synthetase 2 - GrsB1-Acore | | Descriptor: | Gramicidin S synthase 2 | | Authors: | Stephan, P, Basquin, J, Caputi, L, O'Connor, S.E, Kries, H. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Directed Evolution of Piperazic Acid Incorporation by a Nonribosomal Peptide Synthetase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8Q9N
 
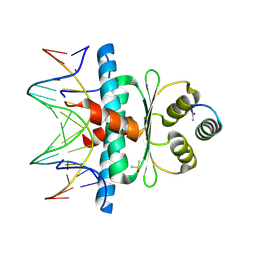 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and MITR deacetylase binding motif mutant L151V. | | Descriptor: | DIMETHYL SULFOXIDE, DNA MADS box, MEF2D protein, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8UWB
 
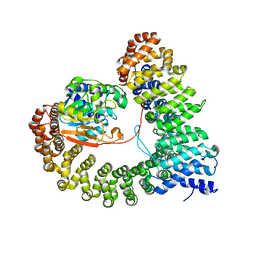 | | Crystal structure of PP2A PPP2R1A-PPP2CA-PPP2R5E phosphatase. | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit epsilon isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wachter, F, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of methylation-independent PP2A assembly guides alphafold2Multimer prediction of family-wide PP2A complexes.
J.Biol.Chem., 300, 2024
|
|
8P06
 
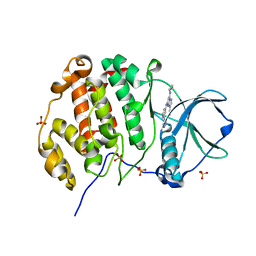 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 5-((2-(4H-1,2,4-triazol-4-yl)pyridin-4-yl)amino)-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidine-3-carbonitrile | | Descriptor: | 7-(cyclopropylamino)-5-[[2-(1,2,4-triazol-4-yl)pyridin-4-yl]amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Brown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | More than an Amide Bioisostere: Discovery of 1,2,4-Triazole-containing Pyrazolo[1,5- a ]pyrimidine Host CSNK2 Inhibitors for Combatting beta-Coronavirus Replication.
J.Med.Chem., 67, 2024
|
|
3K34
 
 | | Human carbonic anhydrase II with a sulfonamide inhibitor | | Descriptor: | (4-SULFAMOYL-PHENYL)-THIOCARBAMIC ACID O-(2-THIOPHEN-3-YL-ETHYL) ESTER, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Behnke, C.A, Le Trong, I, Merritt, E.A, Teller, D.C, Stenkamp, R.E. | | Deposit date: | 2009-10-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic resolution studies of carbonic anhydrase II.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
