2WCF
 
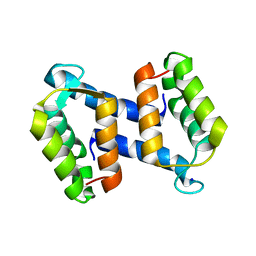 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
5TML
 
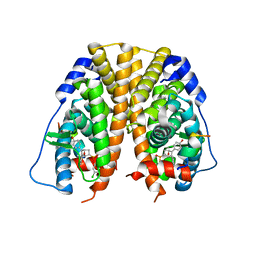 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, (E)-6-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | Descriptor: | 6-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
7BGB
 
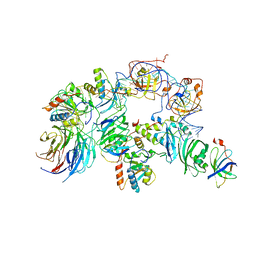 | | The H/ACA RNP lobe of human telomerase | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 2, H/ACA ribonucleoprotein complex subunit 3, ... | | Authors: | Nguyen, T.H.D, Ghanim, G.E, Fountain, A.J, van Roon, A.M.M, Rangan, R, Das, R, Collins, K. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human telomerase holoenzyme with bound telomeric DNA.
Nature, 593, 2021
|
|
6VTK
 
 | |
6VZ4
 
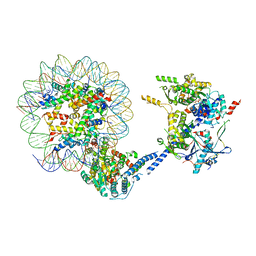 | |
8ANW
 
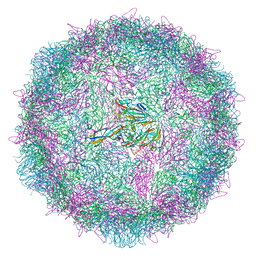 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8). | | Descriptor: | Capsid protein, VP0, VP1, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Production and Characterisation of Stabilised PV-3 Virus-like Particles Using Pichia pastoris .
Viruses, 14, 2022
|
|
4INS
 
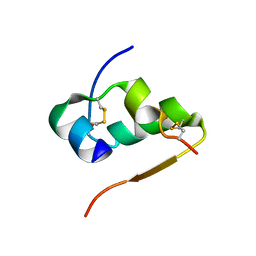 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
6SRS
 
 | | Structure of the Fanconi anaemia core subcomplex | | Descriptor: | Fanconi anaemia protein FANCL, Unassigned secondary structure elements (central region, proposed FANCB-FAAP100), ... | | Authors: | Shakeel, S, Rajendra, E, Alcon, P, He, S, Scheres, S.H.W, Passmore, L.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Fanconi anaemia monoubiquitin ligase complex.
Nature, 575, 2019
|
|
8D9Y
 
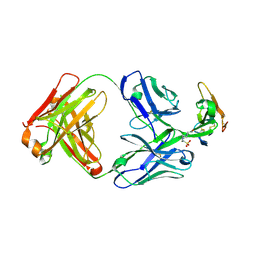 | | Crystal structure of Taipan alpha-neurotoxin in complex with Centi-LNX-D09 antibody | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Centi-LNX-D09 Fab heavy chain, ... | | Authors: | Pletnev, S, Verardi, R, Tully, E.S, Glanville, J, Kwong, P.D. | | Deposit date: | 2022-06-12 | | Release date: | 2023-06-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Venom protection by antibody from a snakebite hyperimmune subject
To Be Published
|
|
6NUO
 
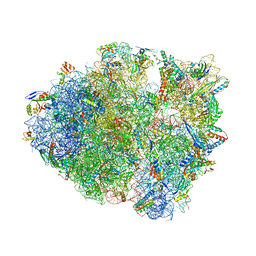 | | Modified tRNA(Pro) bound to Thermus thermophilus 70S (cognate) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-02-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6SD4
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 34-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
8DA0
 
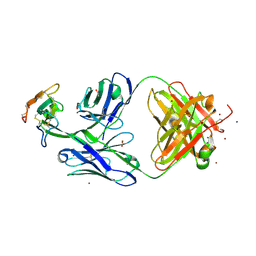 | | Crystal structure of Mamba alpha-neurotoxin in complex with Centi-LNX-D09 antibody | | Descriptor: | ACETATE ION, Alpha-elapitoxin-Dpp2d, Centi-LNX-D09 Fab heavy chain, ... | | Authors: | Pletnev, S, Verardi, R, Tully, E.S, Glanville, J, Kwong, P.D. | | Deposit date: | 2022-06-12 | | Release date: | 2023-06-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Venom protection by antibody from a snakebite hyperimmune subject
To Be Published
|
|
1MUP
 
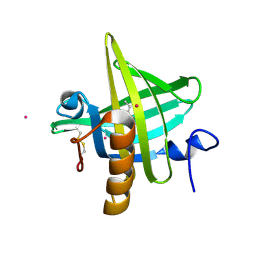 | | PHEROMONE BINDING TO TWO RODENT URINARY PROTEINS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN | | Authors: | Bocskei, Z, Flower, D.R, Groom, C.R, Phillips, S.E.V, North, A.C.T. | | Deposit date: | 1992-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography.
Nature, 360, 1992
|
|
8DFY
 
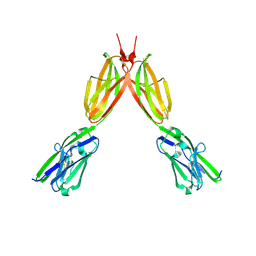 | | Crystal structure of Human BTN2A1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, ... | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.552 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
7ABR
 
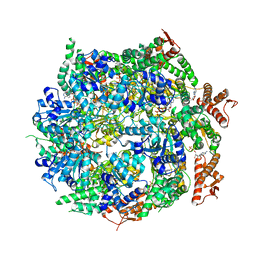 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
6SGA
 
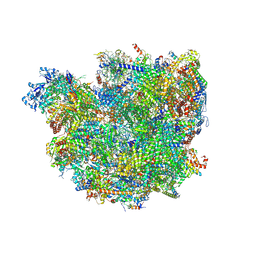 | | Body domain of the mt-SSU assemblosome from Trypanosoma brucei | | Descriptor: | 9S rRNA, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Saurer, M, Ramrath, D.J.F, Niemann, M, Calderaro, S, Prange, C, Mattei, S, Scaiola, A, Leitner, A, Bieri, P, Horn, E.K, Leibundgut, M, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-18 | | Last modified: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery.
Science, 365, 2019
|
|
4DR7
 
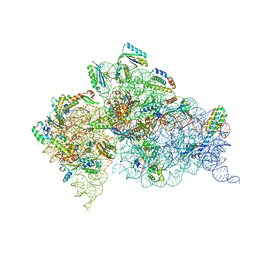 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position, and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DR4
 
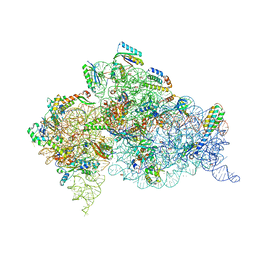 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, cognate transfer RNA anticodon stem-loop and multiple copies of paromomycin molecules bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.969 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
8DAI
 
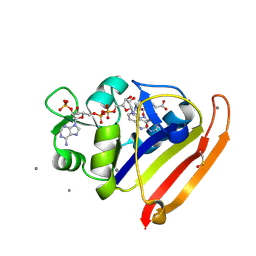 | | E. coli DHFR complex with NADP+ and 10-methylfolate | | Descriptor: | Dihydrofolate reductase, MANGANESE (II) ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Greisman, J.B, Brookner, D.E, Hekstra, D.R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7ADQ
 
 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno-Chicano, T.M, Ebrahim, A.E, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | Deposit date: | 2020-09-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
6TK9
 
 | | Purine-nucleoside phosphorylase from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Timofeev, V.I, Abramchik, Y.A, Kostromina, M.A, Tuzova, E.S, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2019-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Purine-nucleoside phosphorylase from Thermus thermophilus
To Be Published
|
|
8DK4
 
 | | Peroxisome proliferator-activated receptor gamma in complex with VSP-51-2 | | Descriptor: | 5-(benzylcarbamoyl)-1-[(4-chloro-3-fluorophenyl)methyl]-1H-indole-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Ma, L, Zhou, X.E, Suino-Powell, K, Hou, N, Zhou, Z, Luo, J, Xu, H.E, Yi, W. | | Deposit date: | 2022-07-02 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of VSP-51-2 as the Novel and Safe PPAR gamma Modulator: Structure-Based Design, Biological Validation and Crystal Analysis
To Be Published
|
|
1AT0
 
 | | 17-kDA fragment of hedgehog C-terminal autoprocessing domain | | Descriptor: | 17-HEDGEHOG | | Authors: | Hall, T.M.T, Porter, J.A, Young, K.E, Koonin, E.V, Beachy, P.A, Leahy, D.J. | | Deposit date: | 1997-08-15 | | Release date: | 1997-11-12 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins.
Cell(Cambridge,Mass.), 91, 1997
|
|
