6EU2
 
 | | Apo RNA Polymerase III - open conformation (oPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
7RJL
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with SHMT | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, GLYCEROL, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
7RJM
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with ILF3 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, Interleukin enhancer-binding factor 3 | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
5CKY
 
 | | Crystal Structure of the MTERF1 R162A substitution bound to the termination sequence. | | Descriptor: | 5' -D (*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3', 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3', Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
7N33
 
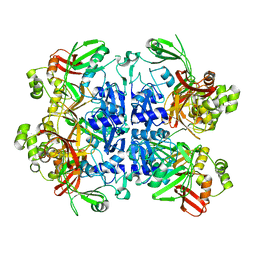 | | SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state | | Descriptor: | RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
6I4E
 
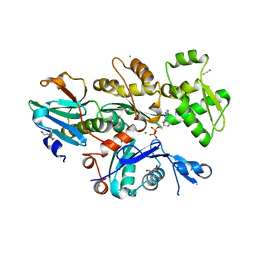 | | Crystal Structure of Plasmodium falciparum actin I in the Mg-ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
5FQL
 
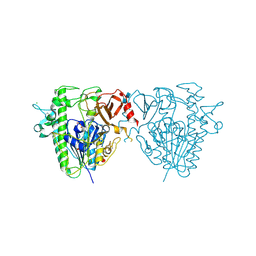 | | Insights into Hunter syndrome from the structure of iduronate-2- sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demydchuk, M, Hill, C.H, Zhou, A, Bunkoczi, G, Stein, P.E, Marchesan, D, Deane, J.E, Read, R.J. | | Deposit date: | 2015-12-11 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into Hunter syndrome from the structure of iduronate-2-sulfatase.
Nat Commun, 8, 2017
|
|
5FUA
 
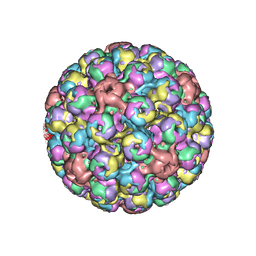 | | Cryo-EM of BK polyomavirus | | Descriptor: | MAJOR CAPSID PROTEIN VP1 | | Authors: | Hurdiss, D.L, Morgan, E.L, Thompson, R.F, Prescott, E.L, Panou, M.M, Macdonald, A, Ranson, N.A. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | New Structural Insights Into the Genome and Minor Capsid Proteins of Bk Polyomavirus Using Cryo-Electron Microscopy.
Structure, 24, 2016
|
|
4WTV
 
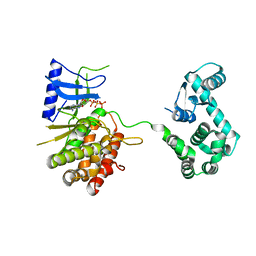 | | Crystal structure of the phosphatidylinositol 4-kinase IIbeta | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-beta,Endolysin,Phosphatidylinositol 4-kinase type 2-beta | | Authors: | Klima, M, Baumlova, A, Chalupska, D, Boura, E. | | Deposit date: | 2014-10-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The high-resolution crystal structure of phosphatidylinositol 4-kinase II beta and the crystal structure of phosphatidylinositol 4-kinase II alpha containing a nucleoside analogue provide a structural basis for isoform-specific inhibitor design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6I5J
 
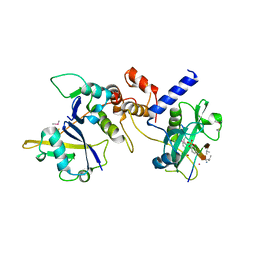 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with growth hormone receptor peptide | | Descriptor: | COBALT (II) ION, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
7RJK
 
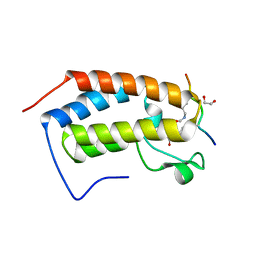 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with hnRNPK | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, GLYCEROL, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
4X2Q
 
 | | Crystal Structure of Human Aldehyde Dehydrogenase, ALDH1a2 | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Retinal dehydrogenase 2 | | Authors: | Stenkamp, R.E, Le Trong, I, Amory, J.K, Paik, J, Goldstein, A.S. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Synthesis and In Vitro Testing of Bisdichloroacetyldiamine Analogs for Use as a Reversible Male Contraceptive
To Be Published
|
|
6MG8
 
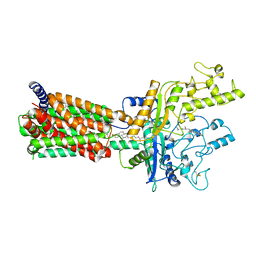 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | Descriptor: | CHOLESTEROL, Protein patched homolog 1 | | Authors: | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
6MA1
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 4a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2R)-2-{[(7-chloro-2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}-2-(2-methoxyphenyl)acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
1US0
 
 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | Authors: | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | Deposit date: | 2003-11-16 | | Release date: | 2004-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.66 Å) | | Cite: | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
7NGO
 
 | |
7QY5
 
 | | Crystal structure of the S.pombe Ars2-Red1 complex. | | Descriptor: | NURS complex subunit pir2, RNA elimination defective protein Red1, ZINC ION | | Authors: | Foucher, A.E, Kadlec, J. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
8QUD
 
 | | Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5 | | Descriptor: | (5R)-5-ethyl-3-(6-spiro[2H-1-benzofuran-3,1'-cyclopropane]-4-yloxypyridin-3-yl)imidazolidine-2,4-dione, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, G, Mckinley, G, Marsden, B, Pike, A.C.W, Ye, M, Brooke, L.M, Bakshi, S, Lakshminarayana, B, Pilati, N, Marasco, A, Gunthorpe, M, Alvaro, G.S, Large, C.H, Williams, E, Sauer, D.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The binding and mechanism of a positive allosteric modulator of Kv3 channels.
Nat Commun, 15, 2024
|
|
7NGX
 
 | |
8BFS
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326 | | Descriptor: | 1,2-ETHANEDIOL, 3~{H}-pyrrolo[2,3-c]isoquinolin-5-amine, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Zhu, W.F, Hernandez-Olmos, V, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326
To Be Published
|
|
7NGK
 
 | |
7NGN
 
 | |
7RJN
 
 | | Crystal structure of human bromodomain containing protein 3 (BRD3) in complex with BCLTF1 | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-associated transcription factor 1, Bromodomain-containing protein 3 | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
6MFS
 
 | | Mouse talin1 residues 1-138 fused to residues 169-400 in complex with phosphatidylinositol 4,5-bisphosphate (PIP2) | | Descriptor: | PHOSPHATE ION, Talin-1 fusion, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Izard, T, Chinthalapudi, K, Rangarajan, E.S. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7NGT
 
 | |
