7ANK
 
 | | Crystal structure of sarcomeric protein FATZ-1 (d91-FATZ-1 construct) in complex with half dimer of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-10-12 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
8EFL
 
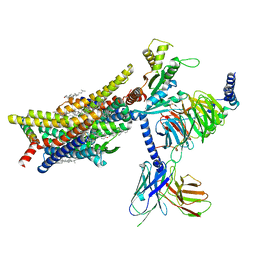 | | SR17018-bound mu-opioid receptor-Gi complex | | Descriptor: | 5,6-dichloro-1-{1-[(4-chlorophenyl)methyl]piperidin-4-yl}-1,3-dihydro-2H-benzimidazol-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
5MQV
 
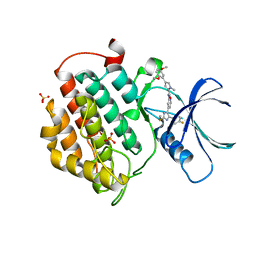 | | Crystal structure of human Casein Kinase I delta in complex with 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide | | Descriptor: | 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
8QYJ
 
 | | Human 20S proteasome assembly structure 1 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome assembly chaperone 3, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EFB
 
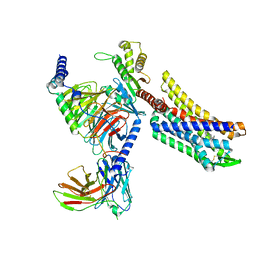 | | Oliceridine-bound mu-opioid receptor-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
5MV1
 
 | | Crystal structure of the E protein of the Japanese encephalitis virulent virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | Descriptor: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
5LN3
 
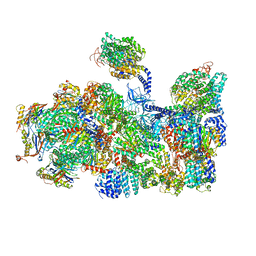 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
7AF8
 
 | | Bacterial 30S ribosomal subunit assembly complex state E (head domain) | | Descriptor: | 16SrRNA (head domain of the 30S ribosome, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
5LRE
 
 | | Crystal structure of Glycogen Phosphorylase b in complex with KS382 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Kyriakis, E, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-31 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase.
Eur J Med Chem, 123, 2016
|
|
6Y0Y
 
 | | Sarcin Ricin Loop, mutant C2666U | | Descriptor: | MAGNESIUM ION, RNA (27-MER), SODIUM ION | | Authors: | Ennifar, E, Westhof, E. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Sarcin Ricin Loop, mutant C2666U
To Be Published
|
|
7Q46
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Pericentriolar material 1 protein | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46002531 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein
To Be Published
|
|
8U48
 
 | | Crystal structure of Bacteroides thetaiotamicron BT1285 D161A-E163A inactive Endoglycosidase in complex with high-mannose N-glycan (Man9GlcNAc2) substrate | | Descriptor: | Endo-beta-N-acetylglucosaminidase, PHOSPHATE ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sastre, D.E, Sultana, N, Navarro, M.V.A.S, Sundberg, E.J. | | Deposit date: | 2023-09-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate.
Nat Commun, 15, 2024
|
|
7Q43
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10 | | Descriptor: | CITRIC ACID, Dedicator of cytokinesis protein 10 peptide, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40002346 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10
To Be Published
|
|
8BCZ
 
 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8QXX
 
 | | HCMV DNA polymerase processivity factor UL44 phosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
4X8A
 
 | | NavMS pore and C-terminal domain grown from protein purified in LiCl | | Descriptor: | HEGA-10, Ion transport protein, NONAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
8QXW
 
 | | HCMV DNA polymerase processivity factor UL44 unphosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
8U46
 
 | |
7Q15
 
 | |
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8U9F
 
 | | Crystal structure of Bacteroides thetaiotamicron BT1285 in complex with NaI | | Descriptor: | 1,2-ETHANEDIOL, Endo-beta-N-acetylglucosaminidase, IODIDE ION, ... | | Authors: | Sastre, D.E, Navarro, M.V.A.S, Sundberg, E.J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate.
Nat Commun, 15, 2024
|
|
7PZ1
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH8535 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-bromanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(3-methoxy-4-methyl-phenyl)piperidine-1-carboxamide, GLYCEROL, ... | | Authors: | Scaletti, E.R, Helleday, T, Stenmark, P. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
