8GRG
 
 | |
8GRH
 
 | | Crystal structure of a constitutively active mutant of the alpha gamma heterodimer of human IDH3 in complex with CIT | | Descriptor: | CITRIC ACID, Human IDH3 alpha subunit, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRU
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ICT, NAD and Ca | | Descriptor: | CALCIUM ION, Human IDH3 alpha subunit, ISOCITRIC ACID, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
3W8I
 
 | |
3W8H
 
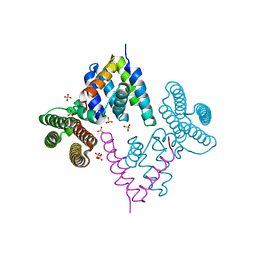 | |
8GS5
 
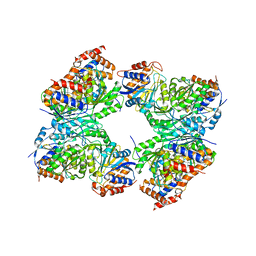 | | Crystal structure of a constitutively active mutant of human IDH3 holoenzyme in apo form | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.486 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GTK
 
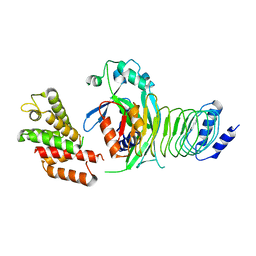 | |
8GTJ
 
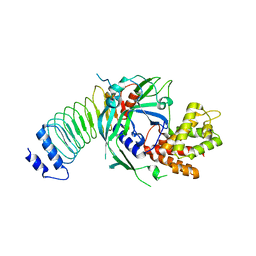 | |
8GTN
 
 | |
4ZB3
 
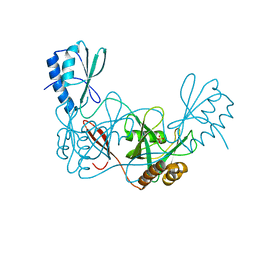 | | Crystal structure of the apo AtNUDT7 | | Descriptor: | Nudix hydrolase 7, SULFATE ION | | Authors: | Tang, Q, Liu, C, Zhong, C, Ding, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Nudix Hydrolase NUDT7 Reveal a Previously Unobserved Conformation.
Mol Plant, 8, 2015
|
|
2F5J
 
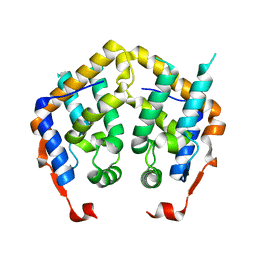 | | Crystal structure of MRG domain from human MRG15 | | Descriptor: | Mortality factor 4-like protein 1 | | Authors: | Zhang, P, Du, J, Ding, J. | | Deposit date: | 2005-11-26 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14
Protein Sci., 15, 2006
|
|
2F5K
 
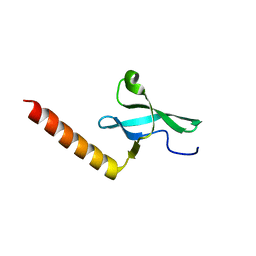 | |
2DSC
 
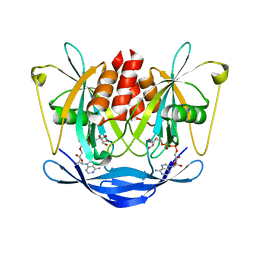 | |
2DSD
 
 | |
2H06
 
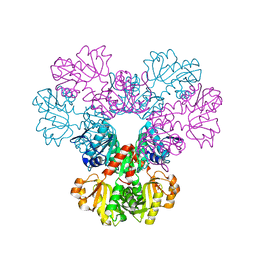 | |
4XPM
 
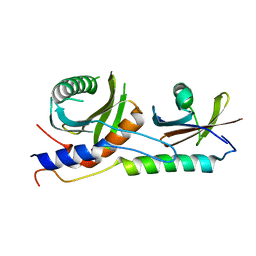 | | Crystal structure of EGO-TC | | Descriptor: | Protein MEH1, Protein SLM4, Uncharacterized protein YCR075W-A | | Authors: | Powis, K, Zhang, T, De Virgilio, C, Ding, J. | | Deposit date: | 2015-01-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling.
Cell Res., 25, 2015
|
|
1O5T
 
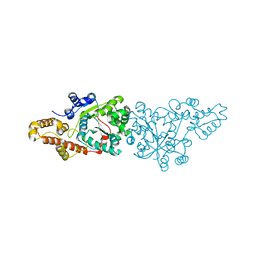 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | Deposit date: | 2003-10-05 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
8H68
 
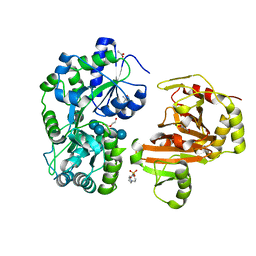 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shi, Y, Ding, J, Yang, H. | | Deposit date: | 2022-10-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
7VKK
 
 | | Crystal structure of D. melanogaster SAMTOR V66W/E67P mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine sensor upstream of mTORC1, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of S -adenosylmethionine sensing by SAMTOR in mTORC1 signaling.
Sci Adv, 8, 2022
|
|
7VKR
 
 | |
7VKQ
 
 | |
5H4B
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
1WAV
 
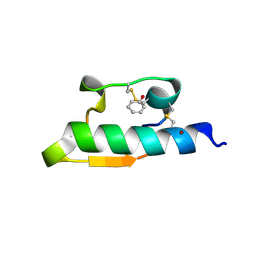 | | CRYSTAL STRUCTURE OF FORM B MONOCLINIC CRYSTAL OF INSULIN | | Descriptor: | INSULIN, PHENOL, ZINC ION | | Authors: | Liang, D.-C, Ding, J.-H, Chang, W.-R, Wan, Z.-L. | | Deposit date: | 1996-02-28 | | Release date: | 1997-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular replacement study on form-B monoclinic crystal of insulin.
Sci.China, Ser.C: Life Sci., 39, 1996
|
|
2DR2
 
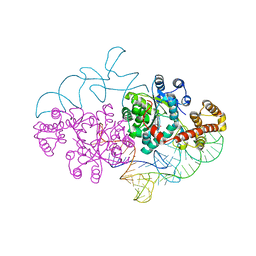 | | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Shen, N, Guo, L, Yang, B, Jin, Y, Ding, J. | | Deposit date: | 2006-06-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) reveals the molecular basis of tRNA recognition and specificity
Nucleic Acids Res., 34, 2006
|
|
