6SI5
 
 | | T. cruzi FPPS in complex with 1-methyl-5-(4,5,6,7-tetrahydrothieno[3,2-c]pyridine-5-carbonyl)pyridin-2(1H)-one | | Descriptor: | 5-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylcarbonyl)-1-methyl-pyridin-2-one, Farnesyl diphosphate synthase, SULFATE ION | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Cornaciu, I, Clavel, D, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
3ICV
 
 | | Structural Consequences of a Circular Permutation on Lipase B from Candida Antartica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
3IFT
 
 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
2MPB
 
 | | NMR structure of BA42 protein from the psychrophilic bacteria Bizionia argentinensis sp. nov | | Descriptor: | BA42 | | Authors: | Cicero, D, Aran, M, Smal, C, Pellizza, L, Gallo, M. | | Deposit date: | 2014-05-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Proteins, 82, 2014
|
|
2N2N
 
 | |
8ARJ
 
 | | Anaplastic Lymphoma Kinase with a novel carboline inhibitor | | Descriptor: | 3-(dimethylamino)-1-[3-[4-(4-methylpiperazin-1-yl)phenyl]-9~{H}-pyrido[2,3-b]indol-6-yl]prop-2-en-1-one, ALK tyrosine kinase receptor | | Authors: | Mologni, L, Scapozza, L, Gambacorti-Passerini, C, Tardy, S, Goekjian, P, Robinson, C, Brown, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Discovery of Novel alpha-Carboline Inhibitors of the Anaplastic Lymphoma Kinase.
Acs Omega, 7, 2022
|
|
2MRY
 
 | |
2MT8
 
 | |
6SHV
 
 | | T. cruzi FPPS in complex with 5-(4-fluorophenoxy)pyridin-2-amine | | Descriptor: | 5-(4-fluoranylphenoxy)pyridin-2-amine, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Cornaciu, I, Clavel, D, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
8AEC
 
 | | Structure of Compound 17 bound to CK2alpha | | Descriptor: | 2-(5-bromanyl-6-chloranyl-1H-indazol-3-yl)ethanenitrile, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Atkinson, E, Iegre, I, Francis, N, Venkitaraman, A, Spring, D, Hyvonen, M. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
6SIH
 
 | |
2MZN
 
 | |
8AEK
 
 | | Structure of Compound 14 bound to CK2alpha | | Descriptor: | 2-[5-(trifluoromethyl)-1H-indol-3-yl]ethanenitrile, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, Fusco, C, Atkinson, E, Iegre, J, Francis-Newton, N, Venkotaraman, A, Spring, D, Hyvonen, M. | | Deposit date: | 2022-07-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
3DYL
 
 | | human phosphdiesterase 9 substrate complex (ES complex) | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, FORMIC ACID, ... | | Authors: | Liu, S, Mansour, M.N, Dillman, K, Perez, J, Danley, D, Menniti, F. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the catalytic mechanism of human phosphodiesterase 9.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3DY8
 
 | | Human Phosphodiesterase 9 in complex with product 5'-GMP (E+P complex) | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, FORMIC ACID, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Liu, S, Mansour, M.N, Dillman, K, Perez, J, Danley, D, Menniti, F. | | Deposit date: | 2008-07-25 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the catalytic mechanism of human phosphodiesterase 9.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3DYN
 
 | | human phosphodiestrase 9 in complex with cGMP (Zn inhibited) | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Liu, S, Mansour, M.N, Dillman, K, Perez, J, Danley, D, Menniti, F. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the catalytic mechanism of human phosphodiesterase 9.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3ICW
 
 | | Structure of a Circular Permutation on Lipase B from Candida Antartica with Bound Suicide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, PHOSPHATE ION, ... | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S.A, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
6SMS
 
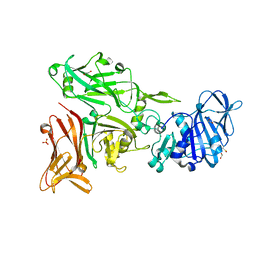 | | Vegetative Insecticidal Protein 1 (Vip1Ac1) from Bacillus thuringiensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Rizkallah, P.J, Al-Maslookhi, H.S, Berry, C, Jones, D. | | Deposit date: | 2019-08-22 | | Release date: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bacillus thuringiensis insecticidal protein 1Ac, VIP1Ac
Not Published
|
|
3FHB
 
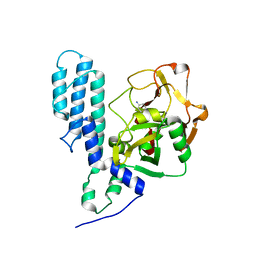 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor 3-aminobenzoic acid | | Descriptor: | 3-AMINOBENZOIC ACID, Poly [ADP-ribose] polymerase 3 | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Schueler, H, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
2MXS
 
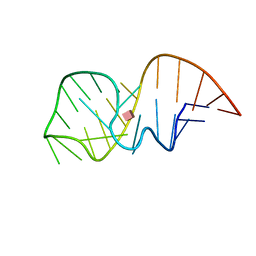 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3IVN
 
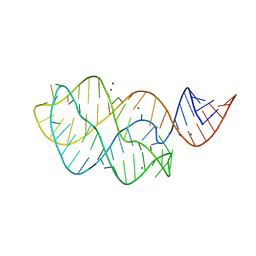 | | Structure of the U65C mutant A-riboswitch aptamer from the Bacillus subtilis pbuE operon | | Descriptor: | A-riboswitch, BROMIDE ION, MAGNESIUM ION | | Authors: | Delfosse, V, Dagenais, P, Chausse, D, Di Tomasso, G, Legault, P. | | Deposit date: | 2009-09-01 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Riboswitch structure: an internal residue mimicking the purine ligand.
Nucleic Acids Res., 38, 2010
|
|
2N0K
 
 | | Chemical shift assignments and structure of the alpha-crystallin domain from human, HSPB5 | | Descriptor: | Alpha-crystallin B chain | | Authors: | Rajagopal, P, Klevit, R.E, Shi, L, Baker, D. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis.
Elife, 4, 2015
|
|
2N8E
 
 | |
2N31
 
 | |
2N86
 
 | | NMR structure of OtTx1a - ICK | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
