7VW5
 
 | |
7VB4
 
 | |
8QUA
 
 | | GTP binding protein YsxC from Staphylococcus aureus | | 分子名称: | ACETYL GROUP, GLYCEROL, Probable GTP-binding protein EngB | | 著者 | Biktimirov, A, Islamov, D, Lazarenko, V, Fatkhullin, B, Validov, S, Yusupov, M, Usachev, K. | | 登録日 | 2023-10-15 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of GTPase YsxC from Staphylococcus aureus.
Biochem.Biophys.Res.Commun., 699, 2024
|
|
5L6V
 
 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a negative allosteric regulator adenosine monophosphate (AMP) - AGPase*AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase, PHOSPHATE ION, ... | | 著者 | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | 登録日 | 2016-05-31 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.667 Å) | | 主引用文献 | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
7VBC
 
 | | Back track state of human RNA Polymerase I Elongation Complex | | 分子名称: | DNA (5'-D(*GP*TP*AP*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*G)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*AP*GP*CP*GP*TP*GP*TP*CP*AP*GP*CP*AP*AP*TP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | 著者 | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | 登録日 | 2021-08-31 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
5KTF
 
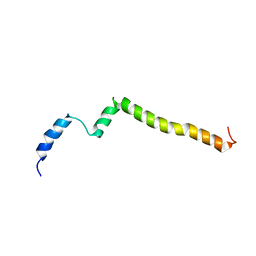 | | Structure of the C-terminal transmembrane domain of scavenger receptor BI (SR-BI) | | 分子名称: | Scavenger receptor class B member 1 | | 著者 | Chadwick, A.C, Peterson, F.C, Volkman, B.F, Sahoo, D. | | 登録日 | 2016-07-11 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the C-Terminal Transmembrane Domain of the HDL Receptor, SR-BI, and a Functionally Relevant Leucine Zipper Motif.
Structure, 25, 2017
|
|
3VX3
 
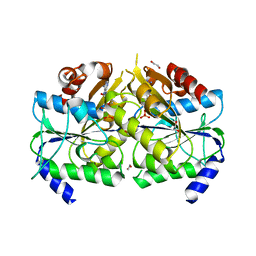 | | Crystal structure of [NiFe] hydrogenase maturation protein HypB from Thermococcus kodakarensis KOD1 | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ... | | 著者 | Sasaki, D, Watanabe, S, Miki, K. | | 登録日 | 2012-09-09 | | 公開日 | 2013-02-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification and Structure of a Novel Archaeal HypB for [NiFe] Hydrogenase Maturation
J.Mol.Biol., 425, 2013
|
|
3VZ0
 
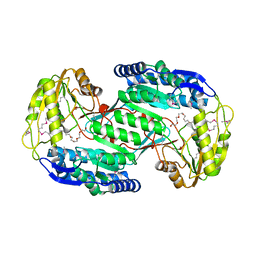 | | Structural insights into cofactor and substrate selection by Gox0499 | | 分子名称: | NONAETHYLENE GLYCOL, Putative NAD-dependent aldehyde dehydrogenase | | 著者 | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | 登録日 | 2012-10-09 | | 公開日 | 2013-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3W0O
 
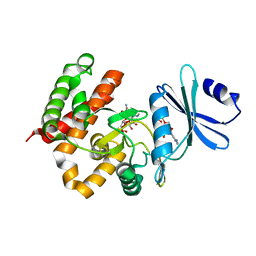 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with ADP and hygromycin B | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | 著者 | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | 登録日 | 2012-11-02 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
5KPE
 
 | | Solution NMR Structure of Denovo Beta Sheet Design Protein, Northeast Structural Genomics Consortium (NESG) Target OR664 | | 分子名称: | De novo Beta Sheet Design Protein OR664 | | 著者 | Tang, Y, Liu, G, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2016-07-03 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
7VBB
 
 | | Structure of the post state human RNA Polymerase I Elongation Complex | | 分子名称: | DNA (25-MER), DNA (5'-D(*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*CP*GP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | 著者 | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | 登録日 | 2021-08-31 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
3NRO
 
 | | Crystal Structure of putative transcriptional factor Lmo1026 from Listeria monocytogenes (FRAGMENT 52-321), Northeast Structural Genomics Consortium Target LmR194 | | 分子名称: | Lmo1026 protein | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-06-30 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target LmR194
To be Published
|
|
8PS0
 
 | |
5LCW
 
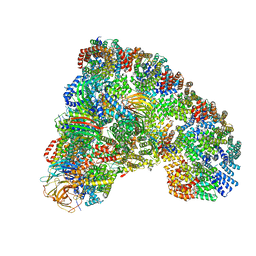 | | Cryo-EM structure of the Anaphase-promoting complex/Cyclosome, in complex with the Mitotic checkpoint complex (APC/C-MCC) at 4.2 angstrom resolution | | 分子名称: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Alfieri, C, Chang, L, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | 登録日 | 2016-06-22 | | 公開日 | 2016-08-10 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular basis of APC/C regulation by the spindle assembly checkpoint.
Nature, 536, 2016
|
|
8PP0
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with JP147 | | 分子名称: | 3-[4-[2,3-dihydro-1H-inden-4-yl(methyl)amino]-6-(trifluoromethyl)pyrimidin-2-yl]oxypropanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | 著者 | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2023-07-05 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Guided Design of a Highly Potent Partial RXR Agonist with Superior Physicochemical Properties.
J.Med.Chem., 67, 2024
|
|
8QAU
 
 | |
5LIE
 
 | |
3W0N
 
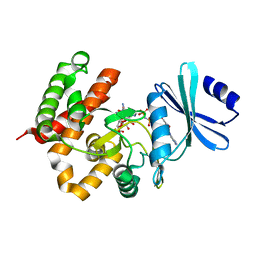 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with AMP-PNP and hygromycin B | | 分子名称: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | 登録日 | 2012-11-02 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
6ALD
 
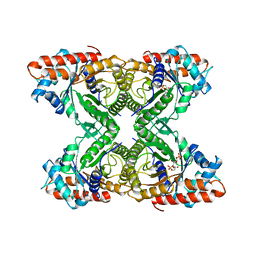 | | RABBIT MUSCLE ALDOLASE A/FRUCTOSE-1,6-BISPHOSPHATE COMPLEX | | 分子名称: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-1,6-BIS(PHOSPHATE) ALDOLASE | | 著者 | Choi, K.H, Mazurkie, A.S, Morris, A.J, Utheza, D, Tolan, D.R, Allen, K.N. | | 登録日 | 1998-12-23 | | 公開日 | 2000-01-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 A(,).
Biochemistry, 38, 1999
|
|
3W15
 
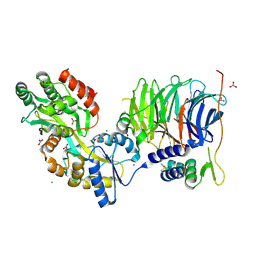 | | Structure of peroxisomal targeting signal 2 (PTS2) of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase in complex with Pex7p and Pex21p | | 分子名称: | 3-ketoacyl-CoA thiolase, peroxisomal, Maltose-binding periplasmic protein, ... | | 著者 | Pan, D, Nakatsu, T, Kato, H. | | 登録日 | 2012-11-06 | | 公開日 | 2013-07-03 | | 最終更新日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Nat.Struct.Mol.Biol., 20, 2013
|
|
7VVE
 
 | | Complex structure of a leaf-branch compost cutinase variant in complex with mono(2-hydroxyethyl) terephthalic acid | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, ... | | 著者 | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | 登録日 | 2021-11-05 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
3VQG
 
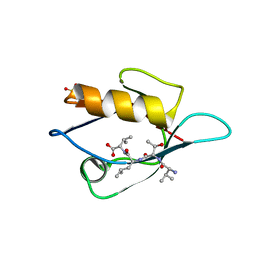 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | 分子名称: | C-terminal peptide from Immunoglobulin superfamily member 5, E3 ubiquitin-protein ligase LNX, SULFATE ION | | 著者 | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | 登録日 | 2012-03-23 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
6AHZ
 
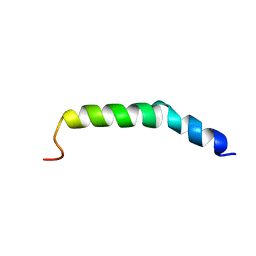 | | The NMR Structure of the Polysialyltranseferase Domain (PSTD) in Polysialyltransferase ST8siaIV | | 分子名称: | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | | 著者 | Liu, X.H, Lu, B, Peng, L.X, Liao, S.M, Zhou, F, Chen, D, Lu, Z.L, Zhou, G.P, Huang, R.B. | | 登録日 | 2018-08-21 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD).
Med Chem, 15, 2019
|
|
8QH4
 
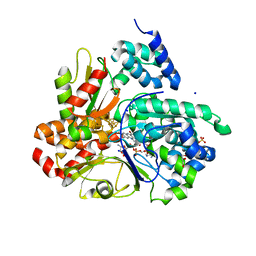 | | Crystal structure of reduced respiratory Complex I subunits NuoEF from Aquifex aeolicus bound to oxidized 3-acetylpyridine adenine dinucleotide | | 分子名称: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Wohlwend, D, Friedrich, T. | | 登録日 | 2023-09-06 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structures of 3-acetylpyridine adenine dinucleotide and ADP-ribose bound to the electron input module of respiratory complex I.
Structure, 32, 2024
|
|
5LFQ
 
 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis (space group P3) | | 分子名称: | Bacterial proteasome activator | | 著者 | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | 登録日 | 2016-07-04 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.503 Å) | | 主引用文献 | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
