1NMQ
 
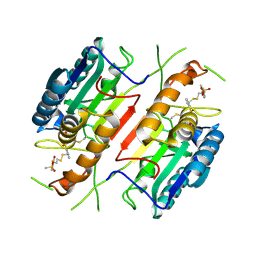 | | Extendend Tethering: In Situ Assembly of Inhibitors | | Descriptor: | 3-(3-{2-[(5-METHANESULFONYL-THIOPHENE-2-CARBONYL)-AMINO]-ETHYLDISULFANYLMETHYL}- BENZENESULFONYLAMINO)-4-OXO-PENTANOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, R.L, DeLano, W, Choong, I.C, Flanagan, M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-10 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
6TGC
 
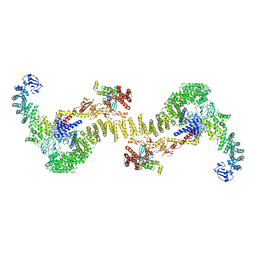 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
7SXN
 
 | | Orb2A residues 1-9 MYNKFVNFI | | Descriptor: | Orb2A residues 1-9 MYNKFVNFI | | Authors: | Bowler, J.T, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2021-11-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Micro-electron diffraction structure of the aggregation-driving N terminus of Drosophila neuronal protein Orb2A reveals amyloid-like beta-sheets.
J.Biol.Chem., 298, 2022
|
|
1HQT
 
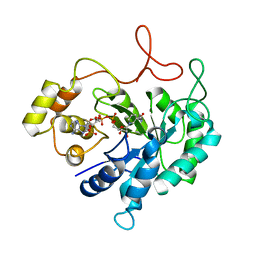 | | THE CRYSTAL STRUCTURE OF AN ALDEHYDE REDUCTASE Y50F MUTANT-NADP COMPLEX AND ITS IMPLICATIONS FOR SUBSTRATE BINDING | | Descriptor: | ALDEHYDE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Hyndman, D, Green, N.C, Li, L, Korithoski, B, Jia, Z, Flynn, T.G. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of an Aldehyde Reductase Y50F Mutant-NADP Complex and its Implications for Substrate Binding
Chem.Biol.Interact., 132, 2001
|
|
3N52
 
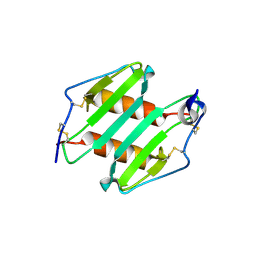 | | crystal Structure analysis of MIP2 | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Rajasekaran, D. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model of GAG/MIP-2/CXCR2 Interfaces and Its Functional Effects.
Biochemistry, 51, 2012
|
|
5DP1
 
 | | Crystal structure of CurK enoyl reductase | | Descriptor: | CurK, GLYCEROL, PHOSPHATE ION | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-12 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
1HIW
 
 | | TRIMERIC HIV-1 MATRIX PROTEIN | | Descriptor: | HIV-1 MATRIX PROTEIN, SULFATE ION | | Authors: | Hill, C.P, Worthylake, D, Bancroft, D.P, Christensen, A.M, Sundquist, W.I. | | Deposit date: | 1996-02-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
5E1X
 
 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 3,4-dichlorophenol bound form | | Descriptor: | 3,4-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
TO BE PUBLISHED
|
|
7TDQ
 
 | |
1HOV
 
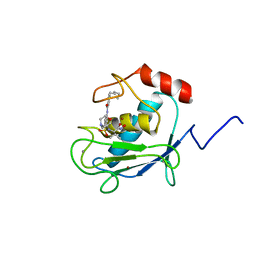 | | SOLUTION STRUCTURE OF A CATALYTIC DOMAIN OF MMP-2 COMPLEXED WITH SC-74020 | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-2, N-{4-[(1-HYDROXYCARBAMOYL-2-METHYL-PROPYL)-(2-MORPHOLIN-4-YL-ETHYL)-SULFAMOYL]-4-PENTYL-BENZAMIDE, ... | | Authors: | Feng, Y, Likos, J.J, Zhu, L, Woodward, H, Munie, G, McDonald, J.J, Stevens, A.M, Howard, C.P, De Crescenzo, G.A, Welsch, D, Shieh, H.-S, Stallings, W.C. | | Deposit date: | 2000-12-11 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the catalytic domain of matrix metalloproteinase-2 complexed with a hydroxamic acid inhibitor
Biochim.Biophys.Acta, 1598, 2002
|
|
1HRF
 
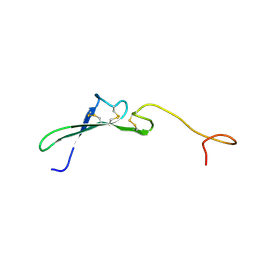 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
6TDG
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 2 | | Descriptor: | 2-chloranyl-3-(4~{H}-1,2,4-triazol-3-yl)aniline, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
1HUJ
 
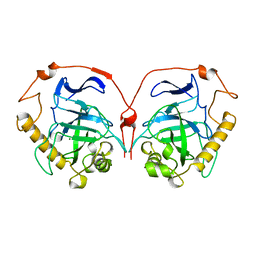 | |
4E0N
 
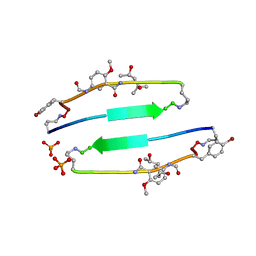 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form II) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5DLC
 
 | |
1MI0
 
 | | Crystal Structure of the redesigned protein G variant NuG2 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Biochemistry, 11, 2002
|
|
1KHE
 
 | | PEPCK complex with nonhydrolyzable GTP analog, MAD data | | Descriptor: | MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Phosphoenolpyruvate Carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1KMO
 
 | | Crystal structure of the Outer Membrane Transporter FecA | | Descriptor: | HEPTANE-1,2,3-TRIOL, Iron(III) dicitrate transport protein fecA, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
3NHC
 
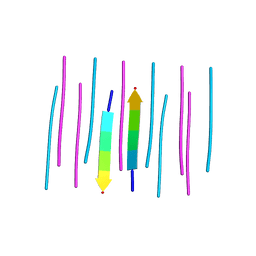 | | GYMLGS segment 127-132 from human prion with M129 | | Descriptor: | Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
5DYI
 
 | | Structure of p97 N-D1 wild-type in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2015-09-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Role of the D1-D2 Linker of Human VCP/p97 in the Asymmetry and ATPase Activity of the D1-domain.
Sci Rep, 6, 2016
|
|
5CSU
 
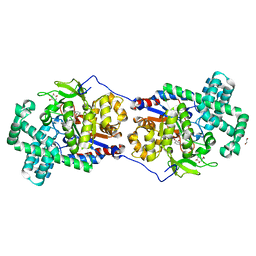 | | Disproportionating enzyme 1 from Arabidopsis - acarviostatin soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
4DN8
 
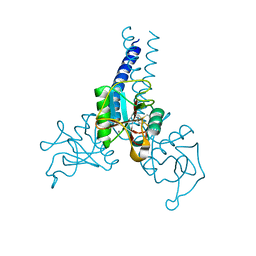 | | Structure of porcine surfactant protein D neck and carbohydrate recognition domain complexed with mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, beta-D-mannopyranose | | Authors: | van Eijk, M, Rynkiewicz, M.J, White, M.R, Hartshorn, K.L, Zou, X, Schulten, K, Luo, D, Crouch, E.C, Cafarella, T.M, Head, J.F, Haagsman, H.P, Seaton, B.A. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Unique Sugar-binding Site Mediates the Distinct Anti-influenza Activity of Pig Surfactant Protein D.
J.Biol.Chem., 287, 2012
|
|
6TPT
 
 | | Crystal structures of FNIII domain three and four of the human leucocyte common antigen-related protein, LAR | | Descriptor: | Receptor-type tyrosine-protein phosphatase F | | Authors: | Vilstrup, J.P, Thirup, S.S, Simonsen, A, Birkefeldt, T, Strandbygaard, D. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal and solution structures of fragments of the human leucocyte common antigen-related protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1KS5
 
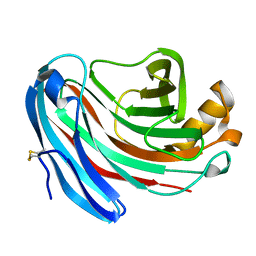 | | Structure of Aspergillus niger endoglucanase | | Descriptor: | Endoglucanase A | | Authors: | Khademi, S, Zhang, D, Swanson, S.M, Wartenberg, A, Witte, C, Meyer, E.F. | | Deposit date: | 2002-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4E0O
 
 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form III) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
