4ZQD
 
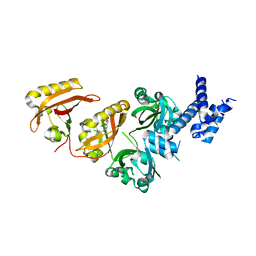 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with the Benzoxadiazole Antagonist 0X3 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
7KP7
 
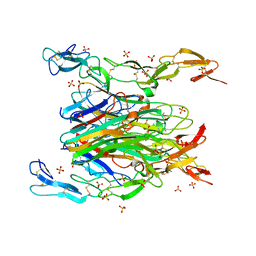 | | asymmetric mTNF-alpha hTNFR1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tumor necrosis factor, ... | | Authors: | Arakaki, T.L, Fox III, D, Edwards, T.E, Foley, A, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the disruption of TNF-TNFR1 signalling by small molecules stabilising a distorted TNF.
Nat Commun, 12, 2021
|
|
4ZIL
 
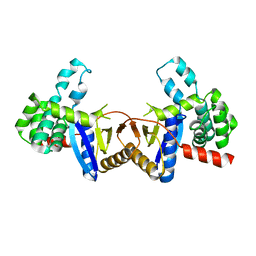 | | Crystal structure of Rv2466c and oxidoreductase from Mycobacterium tuberculosis in its reduced state | | Descriptor: | DSBA oxidoreductase, MAGNESIUM ION | | Authors: | Albesa-Jove, D, Urresti, S, Comino, N, Tersa, M, Guerin, M.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | The Redox State Regulates the Conformation of Rv2466c to Activate the Antitubercular Prodrug TP053.
J.Biol.Chem., 290, 2015
|
|
4ZJW
 
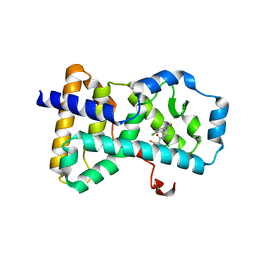 | | RORgamma in complex with inverse agonist 16 | | Descriptor: | 4-chloro-3-[1-(2-chloro-6-fluorobenzoyl)-1,2,3,4-tetrahydroquinolin-6-yl]-N-methylbenzamide, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.J. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of biaryl carboxylamides as potent ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6UCD
 
 | |
6UEN
 
 | |
7KF4
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5IZP
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 10th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-03-25 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
7KMB
 
 | | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
5JMX
 
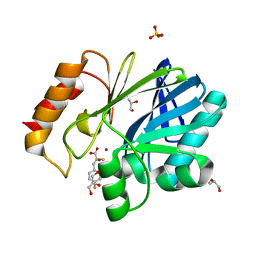 | | Crystal Structure of BcII metallo-beta-lactamase in complex with DZ-305 | | Descriptor: | (2Z)-3-(4-fluorophenyl)-2-sulfanylprop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, Brem, J. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
6IC6
 
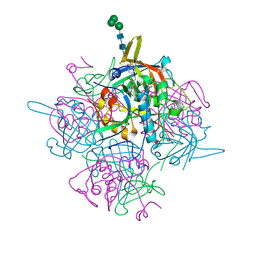 | | Human cathepsin-C in complex with cyclopropyl peptidyl nitrile inhibitor 1 | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{R})-1-(aminomethyl)-2-[4-[4-(trifluoromethyl)phenyl]phenyl]cyclopropyl]-2-azanyl-butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
7KVA
 
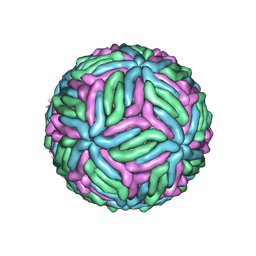 | | Structure of West Nile virus (Kunjin) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Matrix protein M | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7KV9
 
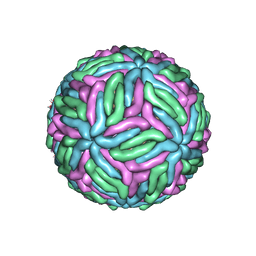 | | Chimeric flavivirus between Binjari virus and West Nile (Kunjin) virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Matrix protein M, envelope protein E | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
8BQ4
 
 | |
7M2N
 
 | | Crystal structure of Human Lactate Dehydrogenase A with Inhibitor Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5-[(5'-{1-(4-carboxy-1,3-thiazol-2-yl)-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-3-yl}-2'-fluoro[1,1'-biphenyl]-4-yl)oxy]-1H-1,2,3-triazole-4-carboxylic acid, ... | | Authors: | Gumpena, R, Ding, J, Powell, D.A, Lowther, W.T. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Glycolate Oxidase/Lactate Dehydrogenase A Inhibitors for Primary Hyperoxaluria
ACS Medicinal Chemistry Letters, 12, 2021
|
|
6BOJ
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN5004 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-{[2-(3-chlorophenyl)-6-ethylpyrimidin-4-yl]methyl}phenyl)acetamide, CHLORIDE ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Memory enhancing effects of BPN14770, an allosteric inhibitor of phosphodiesterase-4D, in wild-type and humanized mice.
Neuropsychopharmacology, 43, 2018
|
|
7M30
 
 | |
7MDW
 
 | |
7M22
 
 | |
6GUQ
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with glucose | | Descriptor: | Putative sugar binding protein, beta-D-glucopyranose | | Authors: | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2018-06-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with glucose
To Be Published
|
|
5FIT
 
 | | FHIT-SUBSTRATE ANALOG | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
6GW6
 
 | |
8P18
 
 | | E167K RF2 on E. coli 70S release complex with UGG (Structure III) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Pundir, S, Larsson, D.S.D, Selmer, M, Sanyal, S. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | The compensatory mechanism of a naturally evolved
E167K RF2 counteracting the loss of
RF1 in bacteria
To Be Published
|
|
6H07
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Nowotny, P, Biggel, P, Schneider, S, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Neutron and X-ray crystal structures of Lactobacillus brevis alcohol dehydrogenase reveal new insights into hydrogen-bonding pathways.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5FTT
 
 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
