2N3V
 
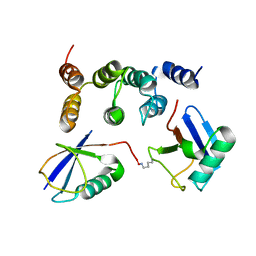 | |
2KQZ
 
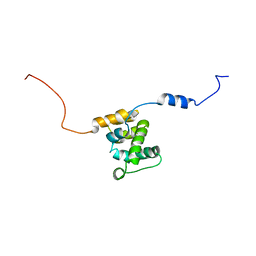 | |
2KR0
 
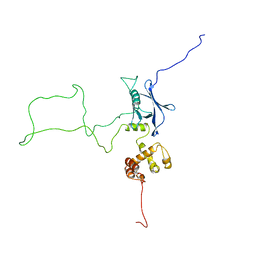 | |
2NBU
 
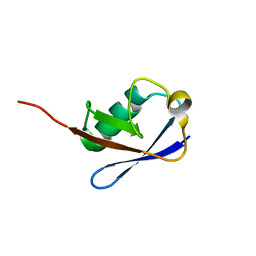 | |
2NBW
 
 | |
2N3U
 
 | |
5XFS
 
 | |
3FUS
 
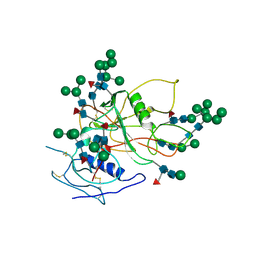 | | Improved Structure of the Unliganded Simian Immunodeficiency Virus gp120 Core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Chen, X, Poon, B, Wang, Q, Ma, J. | | 登録日 | 2009-01-14 | | 公開日 | 2009-06-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5W1X
 
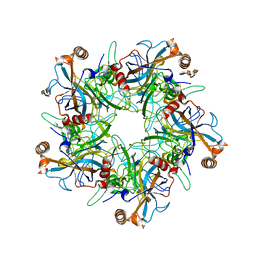 | | Crystal Structure of Humanpapillomavirus18 (HPV18) Capsid L1 Pentamers Bound to Heparin Oligosaccharides | | 分子名称: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, ... | | 著者 | Chen, X.S, Dasgupta, J. | | 登録日 | 2017-06-05 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.374 Å) | | 主引用文献 | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
2NBV
 
 | |
7K17
 
 | |
7K0Y
 
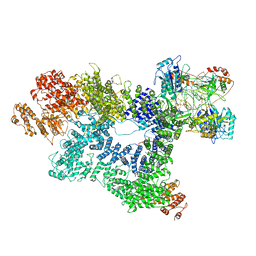 | | Cryo-EM structure of activated-form DNA-PK (complex VI) | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | 著者 | Chen, X, Gellert, M, Yang, W. | | 登録日 | 2020-09-06 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1J
 
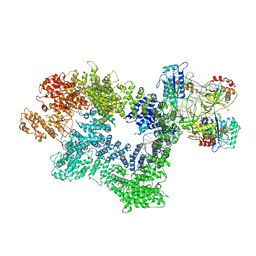 | | CryoEM structure of inactivated-form DNA-PK (Complex III) | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | 著者 | Chen, X, Gellert, M, Yang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1N
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex V) | | 分子名称: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | 著者 | Chen, X, Gellert, M, Yang, W. | | 登録日 | 2020-09-08 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1K
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex IV) | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | 著者 | Chen, X, Gellert, M, Yang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
8JMZ
 
 | | FGFR1 kinase domain with sulfatinib | | 分子名称: | Fibroblast growth factor receptor 1, GLYCEROL, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide, ... | | 著者 | Chen, X.J, Lin, Q.M, Chen, Y.H. | | 登録日 | 2023-06-05 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.988 Å) | | 主引用文献 | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
7V29
 
 | | Crystal structure of FGFR4 with a dual-warhead covalent inhhibitor | | 分子名称: | Fibroblast growth factor receptor 4, N-[2-[[3-(3,5-dimethoxyphenyl)-2-oxidanylidene-1-[3-(4-propanoylpiperazin-1-yl)propyl]-4H-pyrimido[4,5-d]pyrimidin-7-yl]amino]phenyl]propanamide, SULFATE ION | | 著者 | Chen, X.J, Jiang, L.Y, Dai, S.Y, Qu, L.Z, Chen, Y.H. | | 登録日 | 2021-08-07 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.983 Å) | | 主引用文献 | Structure-based design of a dual-warhead covalent inhibitor of FGFR4.
Commun Chem, 5, 2022
|
|
5W77
 
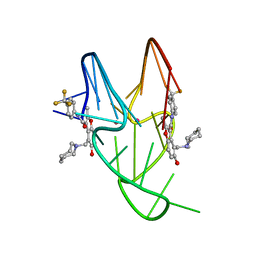 | | Solution structure of the MYC G-quadruplex bound to small molecule DC-34 | | 分子名称: | 4-[(azepan-1-yl)methyl]-5-hydroxy-2-methyl-N-[4-(trifluoromethyl)phenyl]-1-benzofuran-3-carboxamide, DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), POTASSIUM ION | | 著者 | Chen, X, Walters, K.J. | | 登録日 | 2017-06-19 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex.
Nat Commun, 9, 2018
|
|
7K11
 
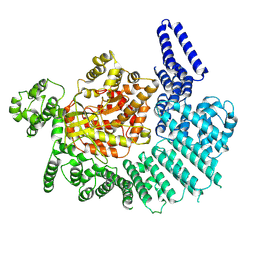 | |
7K10
 
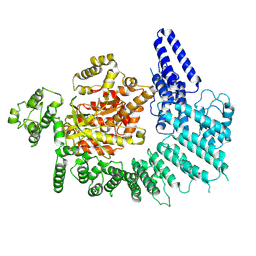 | |
7K19
 
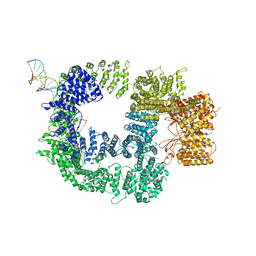 | |
7K1B
 
 | |
5ZXF
 
 | | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a | | 分子名称: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Cheng, X.Y, Su, Z.D, Pi, N, Cao, C.Z, Zhao, Z.T, Zhou, J.J, Chen, R, Kuang, Z.K, Huang, Y.Q. | | 登録日 | 2018-05-19 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a
To Be Published
|
|
6AKO
 
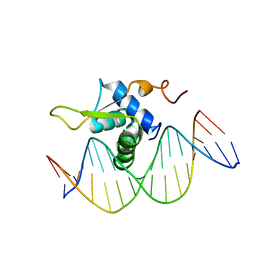 | | Crystal Structure of FOXC2 DBD Bound to DBE2 DNA | | 分子名称: | DNA (5'-D(CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Forkhead box protein C2, ... | | 著者 | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | 登録日 | 2018-09-03 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
6AKP
 
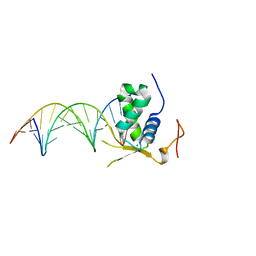 | | Crystal Structural of FOXC2 DNA binding domain bound to PC promoter | | 分子名称: | DNA (5'-D(AP*CP*AP*CP*AP*AP*AP*TP*AP*TP*TP*TP*GP*TP*GP*T)-3'), Forkhead box protein C2, MAGNESIUM ION | | 著者 | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | 登録日 | 2018-09-03 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.323 Å) | | 主引用文献 | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
