8YUU
 
 | | Cryo-EM structure of the histamine-bound H3R-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YUV
 
 | | Cryo-EM structure of the immepip-bound H3R-Gi complex | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
4XDZ
 
 | | Holo structure of ketol-acid reductoisomerase from Ignisphaera aggregans | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2014-12-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
8YUT
 
 | | Cryo-EM structure of the amthamine-bound H2R-Gs complex | | Descriptor: | 5-(2-azanylethyl)-4-methyl-1,3-thiazol-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
1QLY
 
 | | NMR Study of the SH3 Domain From Bruton's Tyrosine Kinase, 20 Structures | | Descriptor: | TYROSINE-PROTEIN KINASE BTK | | Authors: | Tzeng, S.R, Lou, Y.C, Pai, M.T, Chen, C, Chen, S.H, Cheng, J.Y. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Btk SH3 Domain Complexed with a Proline-Rich Peptide from P120Cbl
J.Biomol.NMR, 16, 2000
|
|
6LF4
 
 | | Crystal structure of VMB-1 bound to hydrolyzed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, VMB-1 metallo-beta-lactamase, ZINC ION | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of VMB-1 bound to hydrolyzed meropenem
To Be Published
|
|
6IMQ
 
 | | Crystal structure of PML B1-box multimers | | Descriptor: | CHLORIDE ION, Protein PML, ZINC ION | | Authors: | Li, Y, Ma, X, Chen, Z, Wu, H, Wang, P, Wu, W, Cheng, N, Zeng, L, Zhang, H, Cai, X, Chen, S.J, Chen, Z, Meng, G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis.
Nat Commun, 10, 2019
|
|
7Y72
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface) | | Descriptor: | Fab E7 heavy chain, Fab E7 light chain, Spike glycoprotein | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
7Y71
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab E7 heavy chain, ... | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
4TSK
 
 | | Ketol-acid reductoisomerase from Alicyclobacillus acidocaldarius | | Descriptor: | Ketol-acid reductoisomerase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Uncovering rare NADH-preferring ketol-acid reductoisomerases.
Metab. Eng., 26C, 2014
|
|
4TPO
 
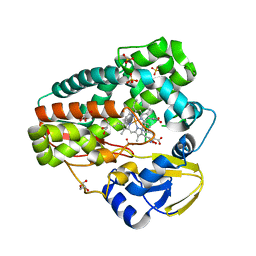 | | High-resolution structure of TxtE with bound tryptophan substrate | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like protein, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Brinkmann-Chen, S, Heinsich, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2014-06-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.225 Å) | | Cite: | Structural, Functional, and Spectroscopic Characterization of the Substrate Scope of the Novel Nitrating Cytochrome P450 TxtE.
Chembiochem, 15, 2014
|
|
4TPN
 
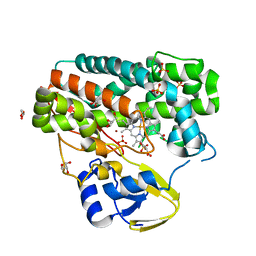 | | High-resolution structure of TxtE in the absence of substrate | | Descriptor: | GLYCEROL, LITHIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Brinkmann-Chen, S, Heinsich, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2014-06-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural, Functional, and Spectroscopic Characterization of the Substrate Scope of the Novel Nitrating Cytochrome P450 TxtE.
Chembiochem, 15, 2014
|
|
3OTW
 
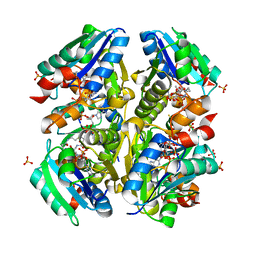 | | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase | | Descriptor: | COENZYME A, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Yin, H.S, Cheng, C.S, Chen, C.G, Luo, Y.C, Chen, W.T, Cheng, S.Y. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase
To be Published
|
|
2MLK
 
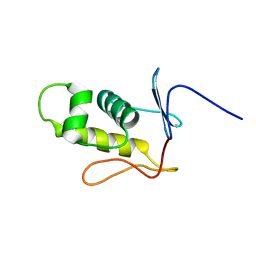 | | Three-dimensional structure of the C-terminal DNA-binding domain of RstA protein from Klebsiella pneumoniae | | Descriptor: | RstA | | Authors: | Fang, P, Chen, S, Cheng, Y, Chang, C, Yu, T, Huang, T. | | Deposit date: | 2014-03-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
5BR4
 
 | | E. coli lactaldehyde reductase (FucO) M185C mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Lactaldehyde reductase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Mutations in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes.
Protein Eng.Des.Sel., 29, 2016
|
|
4KQW
 
 | | The structure of the Slackia exigua KARI in complex with NADP | | Descriptor: | Ketol-acid reductoisomerase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8IW0
 
 | | Crystal structure of the KANK1/liprin-beta1 complex | | Descriptor: | Liprin-beta-1,KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Zhang, J, Chen, S, Wei, Z, Yu, C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
1JXS
 
 | | Solution Structure of the DNA-Binding Domain of Interleukin Enhancer Binding Factor | | Descriptor: | interleukin enhancer binding factor | | Authors: | Chuang, W.J, Liu, P.P, Li, C, Hsieh, Y.H, Chen, S.W, Chen, S.H, Jeng, W.Y. | | Deposit date: | 2001-09-08 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a)
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
4M0W
 
 | | Crystal Structure of SARS-CoV papain-like protease C112S mutant in complex with ubiquitin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Replicase polyprotein 1a, ... | | Authors: | Chou, C.-Y, Chen, H.-Y, Lai, H.-Y, Cheng, S.-C, Chou, Y.-W. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7LSY
 
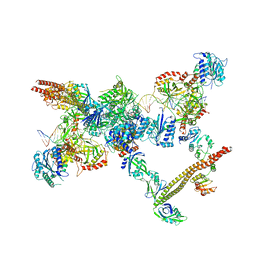 | | NHEJ Short-range synaptic complex | | Descriptor: | DNA (26-MER), DNA (5'-D(P*CP*AP*AP*TP*GP*AP*AP*AP*CP*GP*GP*AP*AP*CP*AP*GP*TP*CP*AP*G)-3'), DNA (5'-D(P*GP*TP*TP*CP*TP*TP*AP*GP*TP*AP*TP*AP*TP*A)-3'), ... | | Authors: | He, Y, Chen, S. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
6J2O
 
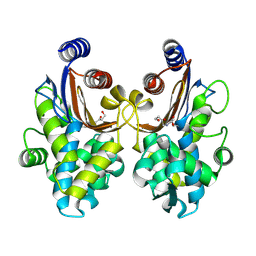 | | Crystal structure of CTX-M-64 clavulanic acid complex | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J25
 
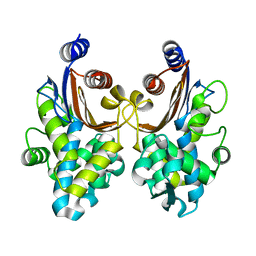 | | CTX-M-64 beta-lactamase mutant-S130T | | Descriptor: | Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-12-30 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
6J2K
 
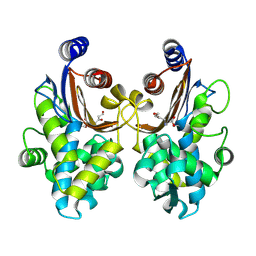 | | CTX-M-64 beta-lactamase S130T clavulanic acid complex | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
4KQX
 
 | | Mutant Slackia exigua KARI DDV in complex with NAD and an inhibitor | | Descriptor: | HISTIDINE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
