3III
 
 | | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Osinski, T, Chruszcz, M, Domagalski, M.J, Cymborowski, M, Shumilin, I.A, Skarina, T, Onopriyenko, O, Zimmerman, M.D, Savchenko, A, Edwards, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-01 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus
To be Published
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
3I7M
 
 | | N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis. | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-08 | | Release date: | 2009-07-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis.
To be Published
|
|
4HUS
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with virginiamycin M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
5FBT
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin | | Descriptor: | CHLORIDE ION, Phosphoenolpyruvate synthase, Rifampin | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
6RZ7
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (F222 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(2-chloranyl-5-fluoranyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3- dihydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6RZ8
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
5FBS
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with ADP and magnesium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoenolpyruvate synthase | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
6RZ5
 
 | | XFEL crystal structure of the human cysteinyl leukotriene receptor 1 in complex with zafirlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
4I62
 
 | | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein AbpA from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Stogios, P.J, Kudritska, M, Wawrzak, Z, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine
To be Published
|
|
4HUR
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with acetyl coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
5IBZ
 
 | | Crystal structure of a novel cyclase (pfam04199). | | Descriptor: | ACETYLPHOSPHATE, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Brown, G, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2016-02-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Crystal structure of a novel cyclase (pfam04199).
To Be Published
|
|
6P2K
 
 | | Crystal structure of AFV00434, an ancestral GH74 enzyme | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
3K6C
 
 | | Crystal structure of protein ne0167 from nitrosomonas europaea | | Descriptor: | Uncharacterized protein NE0167 | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Ne0167 from Nitrosomonas Europaea
To be Published
|
|
6P2O
 
 | | Crystal structure of Streptomyces rapamycinicus GH74 in complex with xyloglucan fragments XLLG and XXXG | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
6PI9
 
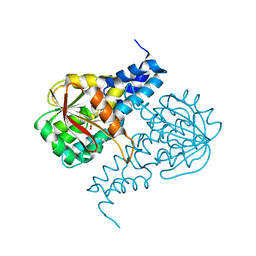 | | Crystal structure of 16S rRNA methyltransferase RmtF in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Stogios, P.J, Kim, Y, Evdokimova, E, Di Leo, R, Semper, C, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 16S rRNA methylase RmtF in complex with S-Adenosyl-L-homocysteine
To be Published
|
|
4H59
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to Bis-tris propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to Bis-tris propane
To be Published
|
|
6B3N
 
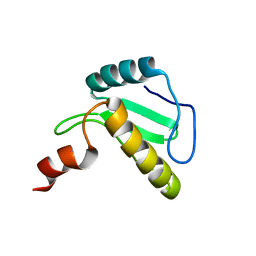 | | Solution structure of the N-terminal domain of the effector NleG5-1 from Escherichia coli O157:H7 str. Sakai | | Descriptor: | NleG5-1 | | Authors: | Valleau, D, Houliston, S, Lemak, A, Anderson, W.F, Arrowsmith, C, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of the effector NleG5-1 from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
6P2L
 
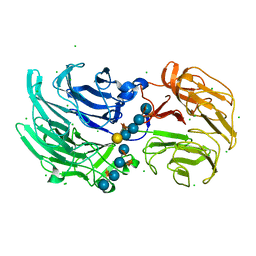 | | Crystal structure of Niastella koreensis GH74 (NkGH74) enzyme | | Descriptor: | CHLORIDE ION, Glycosyl hydrolase BNR repeat-containing protein, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
3K3S
 
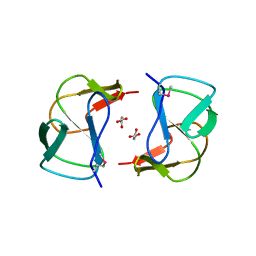 | | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri. | | Descriptor: | ACETATE ION, Altronate hydrolase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Xu, X, Le, B, Zimmerman, M.D, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-04 | | Release date: | 2009-10-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri.
To be Published
|
|
6BNC
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant di-zinc and PEG complex | | Descriptor: | CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
6C5C
 
 | | Crystal structure of the 3-dehydroquinate synthase (DHQS) domain of Aro1 from Candida albicans SC5314 in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, CHLORIDE ION, ... | | Authors: | Michalska, K, Evdokimova, E, Di Leo, R, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-16 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
4JCC
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiuA from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Iron-compound ABC transporter, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.133 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiuA from Streptococcus pneumoniae Canada MDR_19A
TO BE PUBLISHED
|
|
6RZ4
 
 | | Crystal structure of cysteinyl leukotriene receptor 1 in complex with pranlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
