6P9U
 
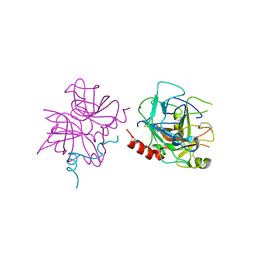 | | Crystal structure of human thrombin mutant W215A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, ZINC ION | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Di Cera, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
6M5T
 
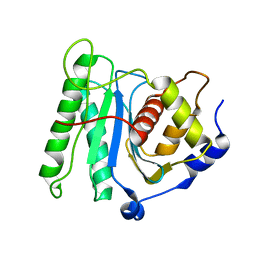 | | The coordinate of the nuclease domain of the apo terminase complex | | Descriptor: | Tripartite terminase subunit 3 | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5R
 
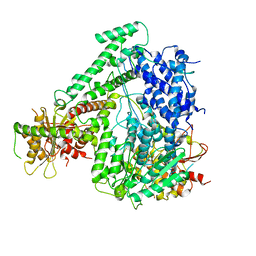 | | The coordinates of the apo monomeric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
7YDW
 
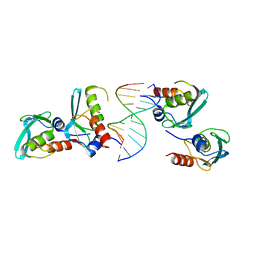 | | Crystal structure of the MPND-DNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MPN domain-containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
7YDT
 
 | | Crystal structure of mouse MPND | | Descriptor: | MPN domain containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
6M5V
 
 | | The coordinate of the hexameric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6V5T
 
 | |
6V64
 
 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
6VLS
 
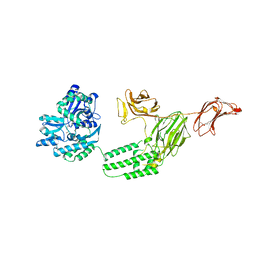 | | Structure of C-terminal fragment of Vip3A toxin | | Descriptor: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Vip3Aa | | Authors: | Jiang, K, Zhang, Y, Chen, Z, Gao, X. | | Deposit date: | 2020-01-25 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Insights into the C-terminal Fragment of Insecticidal Vip3A Toxin ofBacillus thuringiensis.
Toxins, 12, 2020
|
|
6A59
 
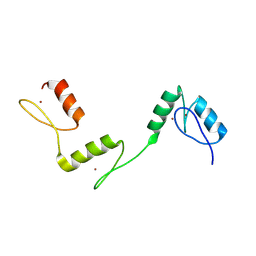 | | Structure of histone demethylase REF6 at 1.8A | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
7Y9I
 
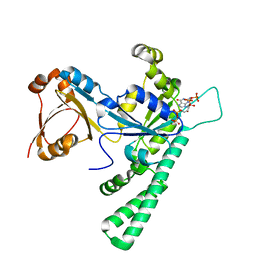 | | Complex structure of AtYchF1 with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Co-crystalization reveals the interaction between AtYchF1 and ppGpp.
Front Mol Biosci, 9, 2022
|
|
8SNX
 
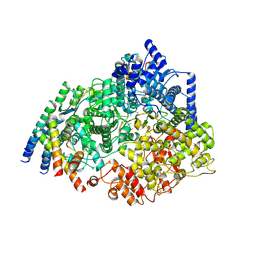 | | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the leader promoter | | Descriptor: | Phosphoprotein, RNA (5'-R(*UP*UP*UP*UP*UP*CP*GP*CP*GP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, D, Gao, Y, Chen, Z, Gooneratne, I, Roesler, C, Mera, C, Liang, B. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the promoter-bound respiratory syncytial virus polymerase.
Nature, 625, 2024
|
|
8SNY
 
 | | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the trailer complementary promoter | | Descriptor: | Phosphoprotein, RNA (5'-R(*UP*UP*UP*UP*UP*CP*UP*CP*GP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, D, Gao, Y, Chen, Z, Gooneratne, I, Roesler, C, Mera, C, Liang, B. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structures of the promoter-bound respiratory syncytial virus polymerase.
Nature, 625, 2024
|
|
8OIO
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Kelch-like protein 12, ... | | Authors: | Dalietou, E.V, Chen, Z, Ramdass, A.E, Manning, C, Richardson, W, Aitmakhanova, K, Platt, M, Pike, A.C.W, Fedorov, O, Brennan, P, Bullock, A.N. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide
To Be Published
|
|
8STL
 
 | | Crystal Structure of Nanobody PIK3_Nb16 against wild-type PI3Kalpha | | Descriptor: | Nanobody PIK3_Nb16, SULFATE ION | | Authors: | Nwafor, J.N, Srinivasan, L, Chen, Z, Gabelli, S.B, Iheanacho, A, Alzogaray, V, Klinke, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of isoform specific nanobodies for Class I PI3Ks
To be published
|
|
7W8L
 
 | | Crystal Structure of Co-type nitrile hydratase mutant from Pseudonocardia thermophila - M46R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
7W8M
 
 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - A129R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
6GIP
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 2, 5-dimethyl core. | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dimethyl-6-quinolin-4-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mathea, S, Chen, Z, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
7YL2
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
6CQS
 
 | | Sediminispirochaeta smaragdinae SPS-1 metallo-beta-lactamase | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Page, R.C, VanPelt, J, Cheng, Z, Crowder, M.W. | | Deposit date: | 2018-03-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Noncanonical Metal Center Drives the Activity of the Sediminispirochaeta smaragdinae Metallo-beta-lactamase SPS-1.
Biochemistry, 57, 2018
|
|
6PXJ
 
 | | Crystal structure of human thrombin mutant I16T | | Descriptor: | GLYCEROL, MAGNESIUM ION, Thrombin heavy chain, ... | | Authors: | Stojanovski, B, Chen, Z, Koester, S.K, Pelc, L.A, Di Cera, E. | | Deposit date: | 2019-07-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the I16-D194 ionic interaction in the trypsin fold.
Sci Rep, 9, 2019
|
|
6PX5
 
 | | CRYSTAL STRUCTURE OF HUMAN MEIZOTHROMBIN DESF1 MUTANT S195A bound with PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Prothrombin, ... | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Gistover, N, Di Cera, E. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
6PXQ
 
 | | Crystal structure of human thrombin mutant D194A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain | | Authors: | Stojanovski, B, Chen, Z, Koester, S.K, Pelc, L.A, Di Cera, E. | | Deposit date: | 2019-07-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of the I16-D194 ionic interaction in the trypsin fold.
Sci Rep, 9, 2019
|
|
1YIQ
 
 | | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADHIIG from Pseudomonas putida HK5. Compariison to the other quinohemoprotein alcohol dehydrogenase ADHIIB found in the same microorganism. | | Descriptor: | CALCIUM ION, HEME C, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Toyama, H, Chen, Z.W, Fukumoto, M, Adachi, O, Matsushita, K, Mathews, F.S. | | Deposit date: | 2005-01-12 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADH-IIG from Pseudomonas putida HK5
J.Mol.Biol., 352, 2005
|
|
6MDX
 
 | | Mechanism of protease dependent DPC repair | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DNA (5'-D(P*CP*C)-3'), ... | | Authors: | Li, F, Raczynska, J, Chen, Z, Yu, H. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-10 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insight into DNA-Dependent Activation of Human Metalloprotease Spartan.
Cell Rep, 26, 2019
|
|
