7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
6LHT
 
 | | Localized reconstruction of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, heavy chain variable region of Fab 18A7, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6DRZ
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N-[(2S)-1-hydroxybutan-2-yl]-1,6-dimethyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRY
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRX
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, BRIL chimera, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DS0
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (1S)-1-[(2-chloro-3,4-dimethoxyphenyl)methyl]-6-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5TVN
 
 | | Crystal structure of the LSD-bound 5-HT2B receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, S, McCorvy, J.D, Betz, R.M, Venkatakrishnan, A.J, Levit, A, Lansu, K, Schools, Z.L, Che, T, Nichols, D.E, Shoichet, B.K, Dror, R.O, Roth, B.L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of an LSD-Bound Human Serotonin Receptor.
Cell, 168, 2017
|
|
6EBV
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with ADP and pantothenate analog Deoxy-N7-Pan | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N-heptyl-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, Type II pantothenate kinase | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H, Park, H. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
6BGJ
 
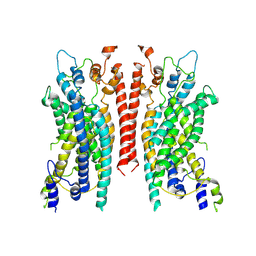 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
5LB2
 
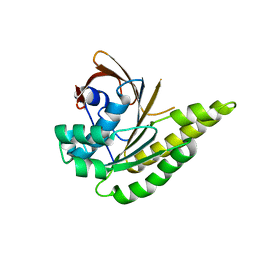 | | Apo-structure of humanised RadA-mutant humRadA2 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
7MLK
 
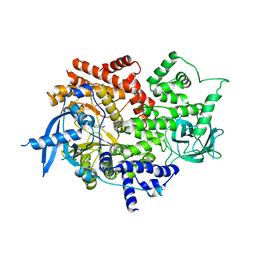 | | Crystal structure of human PI3Ka (p110a subunit) with MMV085400 bound to the active site determined at 2.9 angstroms resolution | | Descriptor: | 4-[6-(3,4,5-trimethoxyanilino)pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Krake, S.H, Martinez, P.D.G, Poggi, M.L, Ferreira, M.S, Aguiar, A.C.C, Souza, G.E, Wenlock, M, Jones, B, Steinbrecher, T, Day, T, McPhail, J, Burke, J, Yeo, T, Mok, S, Uhlemann, A.C, Fidock, D.A, Chen, P, Grodsky, N, Deng, Y.L, Guido, R.V.C, Campbell, S.F, Willis, P.A, Dias, L.C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazines as potent PI4K inhibitors with antimalarial activity
To Be Published
|
|
5LYW
 
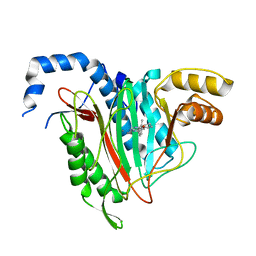 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX; WITH AN INHIBITOR 6-((R)-2-o-Tolyloxymethyl-pyrrolidin-1-yl)-9H-purine | | Descriptor: | 6-[(2~{R})-2-[(2-methylphenoxy)methyl]pyrrolidin-1-yl]-7~{H}-purine, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Musil, D, Heinrich, T, Knoechel, T, Lehmann, M. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Novel reversible methionine aminopeptidase-2 (MetAP-2) inhibitors based on purine and related bicyclic templates.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8D1P
 
 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 7-Deaza-2'-C-methyladenosine | | Descriptor: | 7-(2-C-methyl-beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D1S
 
 | | Crystal structure of Plasmodium falciparum GRP78 in complex with Toyocamycin | | Descriptor: | 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D1Q
 
 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 8-Aminoadenosine | | Descriptor: | (2R,3R,4S,5R)-2-(6,8-diaminopurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
5N2M
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, propan-2-yl ~{N}-[(2~{S},4~{R})-6-(3-acetamidophenyl)-1-ethanoyl-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
5N2L
 
 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, SULFATE ION, ... | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
6AWJ
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with ADP and pantothenate analog Deoxy-MeO-N5Pan with pantothenate present in reaction | | Descriptor: | (2R)-2-hydroxy-N-{3-[(5-methoxypentyl)amino]-3-oxopropyl}-3,3-dimethylbutanamide, ADENOSINE-5'-DIPHOSPHATE, Type II pantothenate kinase | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
6AWI
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with ADP and pantothenate analog Deoxy-N5Pan | | Descriptor: | (2R)-2-hydroxy-3,3-dimethyl-N-[3-oxo-3-(pentylamino)propyl]butanamide, ADENOSINE-5'-DIPHOSPHATE, Type II pantothenate kinase | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
6AWH
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with ATP and pantothenate analog Deoxy-MeO-N5Pan | | Descriptor: | (2R)-2-hydroxy-N-{3-[(5-methoxypentyl)amino]-3-oxopropyl}-3,3-dimethylbutanamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
6AVP
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with ADP and pantothenate analog Phosphate-MeO-N5Pan | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(5-methoxypentyl)-beta-alaninamide, ... | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
6AWG
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with nucleotides and pantothenate analog Deoxy-N190Pan | | Descriptor: | (2R)-N-(3-{[(2H-1,3-benzodioxol-5-yl)methyl]amino}-3-oxopropyl)-2-hydroxy-3,3-dimethylbutanamide, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
6DFO
 
 | | Crystal structure of human GRP78 in complex with 8-bromoadenosine | | Descriptor: | 8-bromoadenosine, Endoplasmic reticulum chaperone BiP | | Authors: | Ferrie, R.P, Chen, Y, Antoshchenko, T, Cooney, O.M, Huang, Y, Park, H.W. | | Deposit date: | 2018-05-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of human GRP78 in complex with 8-bromoadenosine
To be Published
|
|
8SZV
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog SJPYT-318 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-[(4-phenoxyphenyl)methyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Li, Y, Chen, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand flexibility and binding pocket malleability cooperate to allow selective PXR activation by analogs of a promiscuous nuclear receptor ligand.
Structure, 31, 2023
|
|
