6ITY
 
 | | CTX-M-64 sulbactam complex | | Descriptor: | ACRYLIC ACID, Beta-lactamase, TRANS-ENAMINE INTERMEDIATE OF SULBACTAM | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-11-26 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
7EPW
 
 | | Crystal structure of monooxygenase Tet(X4) with tigecycline | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, TIGECYCLINE | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2021-04-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline.
Bmc Biol., 19, 2021
|
|
7EPV
 
 | | Crystal structure of tigecycline degrading monooxygenase Tet(X4) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, GLYCEROL | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline.
Bmc Biol., 19, 2021
|
|
4TPO
 
 | | High-resolution structure of TxtE with bound tryptophan substrate | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like protein, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Brinkmann-Chen, S, Heinsich, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2014-06-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.225 Å) | | Cite: | Structural, Functional, and Spectroscopic Characterization of the Substrate Scope of the Novel Nitrating Cytochrome P450 TxtE.
Chembiochem, 15, 2014
|
|
4TPN
 
 | | High-resolution structure of TxtE in the absence of substrate | | Descriptor: | GLYCEROL, LITHIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Brinkmann-Chen, S, Heinsich, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2014-06-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural, Functional, and Spectroscopic Characterization of the Substrate Scope of the Novel Nitrating Cytochrome P450 TxtE.
Chembiochem, 15, 2014
|
|
4ML3
 
 | | X-ray structure of ComE D58A REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
5BYZ
 
 | | ERK5 in complex with small molecule | | Descriptor: | 4-({5-fluoro-4-[2-methyl-1-(propan-2-yl)-1H-imidazol-5-yl]pyrimidin-2-yl}amino)-N-[2-(piperidin-1-yl)ethyl]benzamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4MLD
 
 | | X-ray structure of ComE D58E REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
5BYY
 
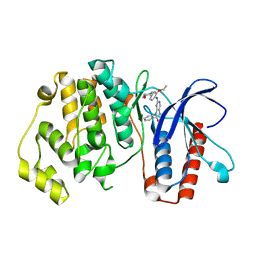 | | ERK5 IN COMPLEX WITH SMALL MOLECULE | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Mitogen-activated protein kinase 7 | | Authors: | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2ZJO
 
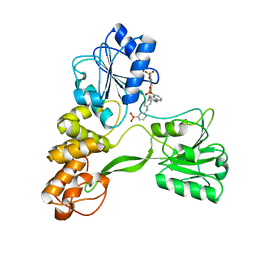 | | Crystal structure of hepatitis C virus NS3 helicase with a novel inhibitor | | Descriptor: | Genome polyprotein, N-[4-(BIS{4-[(3-SULFOPHENYL)AMINO]PHENYL}METHYLENE)CYCLOHEXA-2,5-DIEN-1-YLIDENE]-4-SULFOBENZENAMINIUM | | Authors: | Liaw, S.H, Chen, S.J, Hu, C.Y, Chi, W.K, Chu, I.D, Hwang, L.H, Chern, J.W, Chen, D.S. | | Deposit date: | 2008-03-07 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Discovery of Triphenylmethane Derivatives as Novel Inhibitors of Hepatitis C Virus Helicase
To be Published
|
|
5E4R
 
 | | Crystal structure of domain-duplicated synthetic class II ketol-acid reductoisomerase 2Ia_KARI-DD | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Buller, A.R, Arnold, F.H. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Artificial domain duplication replicates evolutionary history of ketol-acid reductoisomerases.
Protein Sci., 25, 2016
|
|
5BR4
 
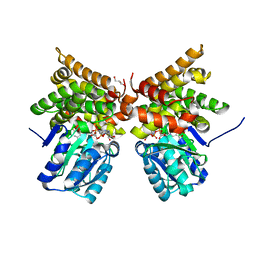 | | E. coli lactaldehyde reductase (FucO) M185C mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Lactaldehyde reductase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Mutations in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes.
Protein Eng.Des.Sel., 29, 2016
|
|
2MLK
 
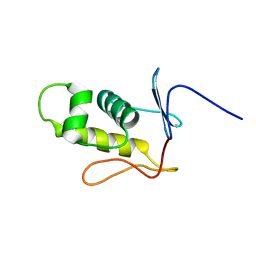 | | Three-dimensional structure of the C-terminal DNA-binding domain of RstA protein from Klebsiella pneumoniae | | Descriptor: | RstA | | Authors: | Fang, P, Chen, S, Cheng, Y, Chang, C, Yu, T, Huang, T. | | Deposit date: | 2014-03-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
3J9K
 
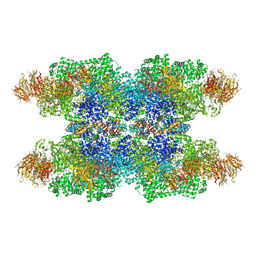 | | Structure of Dark apoptosome in complex with Dronc CARD domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apaf-1 related killer DARK, Caspase Nc | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
4KQW
 
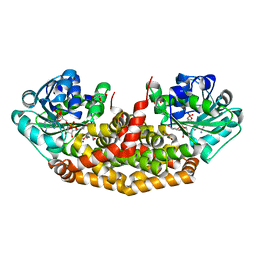 | | The structure of the Slackia exigua KARI in complex with NADP | | Descriptor: | Ketol-acid reductoisomerase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1QLY
 
 | | NMR Study of the SH3 Domain From Bruton's Tyrosine Kinase, 20 Structures | | Descriptor: | TYROSINE-PROTEIN KINASE BTK | | Authors: | Tzeng, S.R, Lou, Y.C, Pai, M.T, Chen, C, Chen, S.H, Cheng, J.Y. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Btk SH3 Domain Complexed with a Proline-Rich Peptide from P120Cbl
J.Biomol.NMR, 16, 2000
|
|
8IW0
 
 | | Crystal structure of the KANK1/liprin-beta1 complex | | Descriptor: | Liprin-beta-1,KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Zhang, J, Chen, S, Wei, Z, Yu, C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
1DXO
 
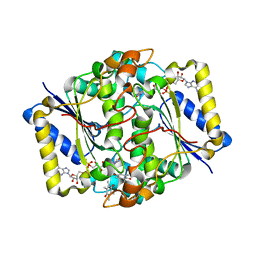 | | Crystal structure of human NAD[P]H-QUINONE oxidoreductase CO with 2,3,5,6,tetramethyl-P-benzoquinone (duroquinone) at 2.5 Angstrom resolution | | Descriptor: | DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-12 | | Release date: | 2000-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DXQ
 
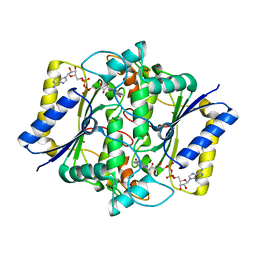 | | CRYSTAL STRUCTURE OF MOUSE NAD[P]H-QUINONE OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1D4A
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AT 1.7 A RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4KQX
 
 | | Mutant Slackia exigua KARI DDV in complex with NAD and an inhibitor | | Descriptor: | HISTIDINE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4M0W
 
 | | Crystal Structure of SARS-CoV papain-like protease C112S mutant in complex with ubiquitin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Replicase polyprotein 1a, ... | | Authors: | Chou, C.-Y, Chen, H.-Y, Lai, H.-Y, Cheng, S.-C, Chou, Y.-W. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8Q96
 
 | | P301S Tau Filaments from the Brains of Tg2541 Transgenic Mouse Line | | Descriptor: | Isoform Tau-E of Microtubule-associated protein tau, Unknown protein | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
8Q92
 
 | | P301S Tau Filaments from the Brains of PS19 Transgenic Mouse Line | | Descriptor: | Microtubule-associated protein tau | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
1H69
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,3,5,6,TETRAMETHYL-P-BENZOQUINONE (DUROQUINONE) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | 3-(HYDROXYMETHYL)-1-METHYL-5-(2-METHYLAZIRIDIN-1-YL)-2-PHENYL-1H-INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-08 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
