6ZH8
 
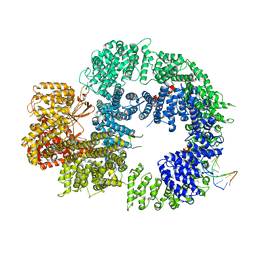 | | Cryo-EM structure of DNA-PKcs:DNA | | 分子名称: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5LQI
 
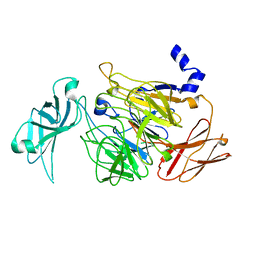 | |
7NFC
 
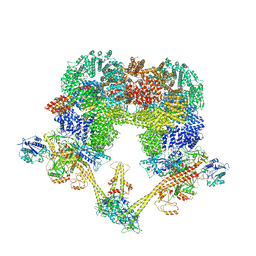 | | Cryo-EM structure of NHEJ super-complex (dimer) | | 分子名称: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2021-02-05 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NFE
 
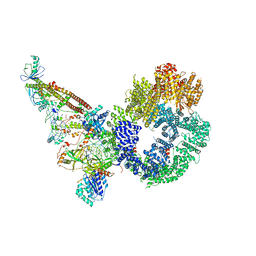 | | Cryo-EM structure of NHEJ super-complex (monomer) | | 分子名称: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2021-02-06 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
4UNM
 
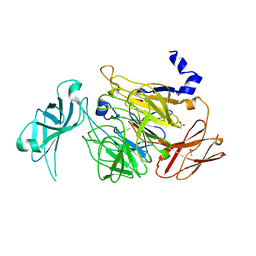 | |
6ZFP
 
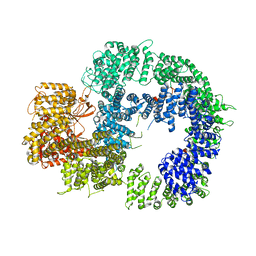 | | Cryo-EM structure of DNA-PKcs (State 2) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
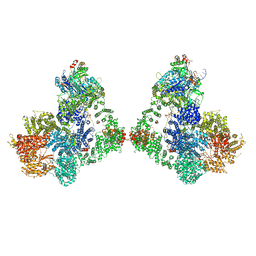 | | Cryo-EM structure of DNA-PK dimer | | 分子名称: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-23 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.24 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
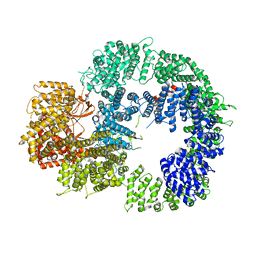 | | Cryo-EM structure of DNA-PKcs (State 1) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
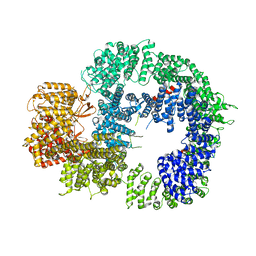 | | Cryo-EM structure of DNA-PKcs (State 3) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
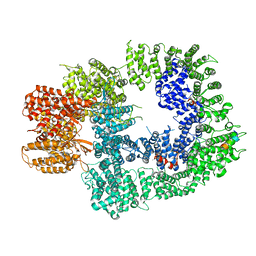 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHA
 
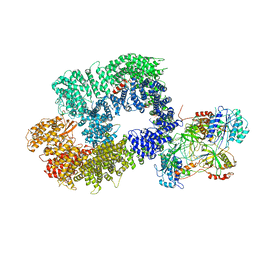 | | Cryo-EM structure of DNA-PK monomer | | 分子名称: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5LXZ
 
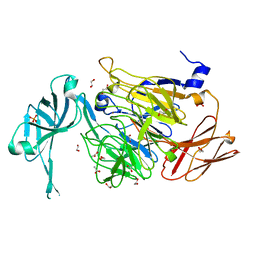 | |
5FTZ
 
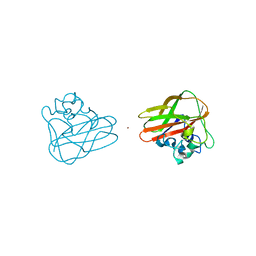 | | AA10 lytic polysaccharide monooxygenase (LPMO) from Streptomyces lividans | | 分子名称: | CHITIN BINDING PROTEIN, COPPER (II) ION | | 著者 | Chaplin, A.K.C, Wilson, M.T, Hough, M.A, Svistunenko, D.A, Hemsworth, G.R, Walton, P.H, Vijgenboom, E, Worrall, J.A.R. | | 登録日 | 2016-01-19 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Heterogeneity in the Histidine-Brace Copper Coordination Sphere in Aa10 Lytic Polysaccharide Monooxygenases.
J.Biol.Chem., 291, 2016
|
|
6ZJI
 
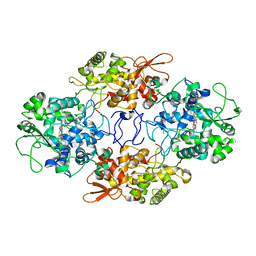 | |
8RWY
 
 | |
6EI0
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: apo form | | 分子名称: | Cytosolic copper storage protein (Ccsp), GLYCEROL, SULFATE ION, ... | | 著者 | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | 登録日 | 2017-09-15 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
6EK9
 
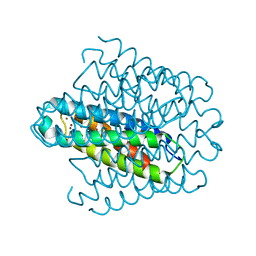 | | Cytosolic copper storage protein Csp from Streptomyces lividans: Cu loaded form | | 分子名称: | COPPER (I) ION, Cytosolic copper storage protein | | 著者 | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | 登録日 | 2017-09-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
8BOT
 
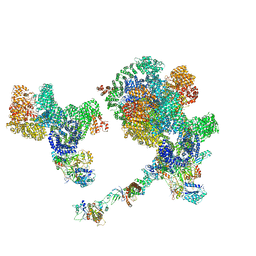 | |
7ZVT
 
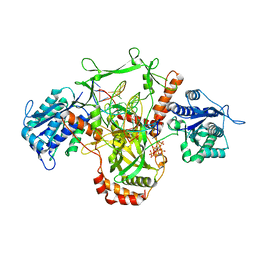 | | CryoEM structure of Ku heterodimer bound to DNA | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZWA
 
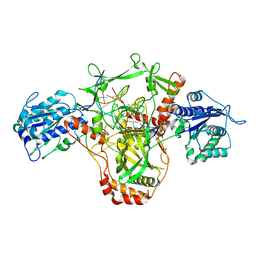 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-19 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
7ZYG
 
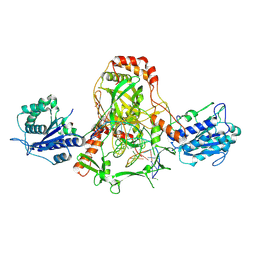 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | 分子名称: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-24 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
8BHY
 
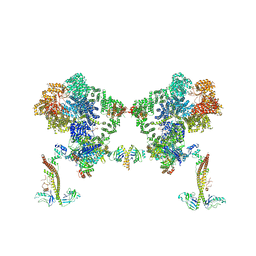 | |
8BH3
 
 | |
8BHV
 
 | |
7ZT6
 
 | |
