7ACD
 
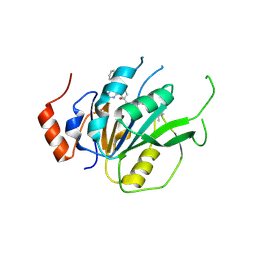 | | Crystal structure of the human METTL3-METTL14 complex with compound T30 (UZH1a) | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-oxidanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes.
Chemmedchem, 16, 2021
|
|
5OWW
 
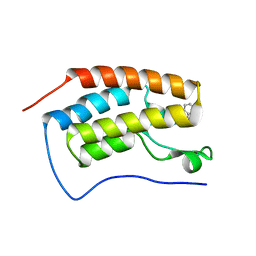 | | Crystal structure of human BRD4(1) bromodomain in complex with UT22B | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(3-methylbenzotriazol-5-yl)-1-(phenylmethyl)imidazole-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-09-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
Acs Cent.Sci., 4, 2018
|
|
5OVB
 
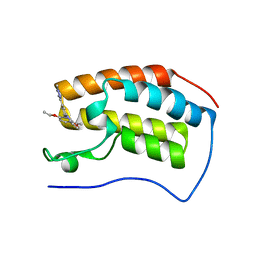 | | Crystal structure of human BRD4(1) bromodomain in complex with DR46 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(1-methylpyrazol-3-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-08-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
Acs Cent.Sci., 4, 2018
|
|
5OWK
 
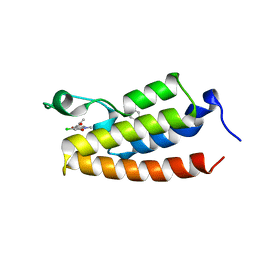 | |
5OWM
 
 | |
6EPT
 
 | | The ATAD2 bromodomain in complex with compound 12 | | Descriptor: | (2~{R})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]piperazine-2-carboxamide, (2~{S})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, ... | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6EPJ
 
 | | The ATAD2 bromodomain in complex with compound 6 | | Descriptor: | (2~{R})-2-azanyl-~{N}-[4-ethanoyl-5-(3-hydroxyphenyl)-1,3-thiazol-2-yl]propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6EPV
 
 | | The ATAD2 bromodomain in complex with compound 5 | | Descriptor: | (2~{R})-2-azanyl-~{N}-(4-oxidanylidene-6,7-dihydro-5~{H}-1,3-benzothiazol-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6EPX
 
 | | The ATAD2 bromodomain in complex with compound 3 | | Descriptor: | (2~{R})-2-carbamimidamido-~{N}-(4-ethanoyl-1,3-thiazolidin-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
5OR9
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 13 | | Descriptor: | (2~{S})-1-(4-fluorophenyl)sulfonyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Vedove, A.D, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
5ORB
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-methoxyphenyl)sulfanyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)ethanamide, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Dalle Vedove, A, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
5OR8
 
 | | Crystal Structure of BAZ2A bromodomain in complex with 1,3-dimethyl-benzimidazolone compound 1 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(4-fluorophenyl)methyl]-1,3,6-trimethyl-2-oxidanylidene-benzimidazole-5-sulfonamide | | Authors: | Lolli, G, Dalle Vedove, A, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
4P4C
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(3-methoxyphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4P5Q
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(2-chlorophenyl)-N-(3-ethoxypropyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4P5Z
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-[4-({[3-(trifluoromethyl)phenyl]carbamoyl}amino)phenyl]-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-20 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
7Z7F
 
 | | Crystal structure of YTHDF2 with compound YLI_DC1_005 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Nai, F, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2022-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7Z7B
 
 | | Crystal structure of YTHDF2 with compound YLI_DC1_003 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Nai, F, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7Z54
 
 | | Crystal structure of YTHDF2 with compound YLI_DC1_006 | | Descriptor: | 9-cyclopropyl-~{N}-methyl-purin-6-amine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nai, F, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
4G2F
 
 | | Human EphA3 kinase domain in complex with compound 7 | | Descriptor: | 1-amino-5-(5-hydroxy-2-methylphenyl)-7,8,9,10-tetrahydropyrimido[4,5-c]isoquinolin-6(5H)-one, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Discovery of a novel chemotype of tyrosine kinase inhibitors by fragment-based docking and molecular dynamics.
ACS MED.CHEM.LETT., 3, 2012
|
|
7B7G
 
 | | BAZ2A bromodomain in complex with compounds MS04 and B11 | | Descriptor: | 1-[1-(3,5-dimethoxyphenyl)piperidin-4-yl]-2,3-dimethyl-guanidine, 5-ethyl-2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazole, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.428 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7B82
 
 | | BAZ2A bromodomain in complex with triazole compound MS04-TN03 | | Descriptor: | (2~{S})-1-[4-[[(~{E})-~{N},~{N}'-dimethylcarbamimidoyl]amino]phenyl]-~{N}-[[2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazol-5-yl]methyl]pyrrolidine-2-carboxamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7B7I
 
 | | BAZ2A bromodomain in complex with triazole compound MS04-TN02 | | Descriptor: | 2-[4-[[(~{E})-~{N},~{N}'-dimethylcarbamimidoyl]amino]piperidin-1-yl]-~{N}-[[2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazol-5-yl]methyl]ethanamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7B7B
 
 | | BAZ2A bromodomain in complex with triazole compound MS04 | | Descriptor: | 5-ethyl-2-(3-methyl-1,2,3-triazol-4-yl)-1~{H}-benzimidazole, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
4GK4
 
 | | Human EphA3 Kinase domain in complex with ligand 90 | | Descriptor: | 8-butyl-1-methyl-7-(5-methyl-1H-indazol-4-yl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
7BC2
 
 | | BAZ2A bromodomain in complex with triazole compound MS04-TN04 | | Descriptor: | 2-(1-methyl-1H-1,2,3-triazol-5-yl)-5-((2-(pyridin-4-yl)pyrrolidin-1-yl)methyl)-1H-benzo[d]imidazole, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Sedykh, M, Caflisch, A, Lolli, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
